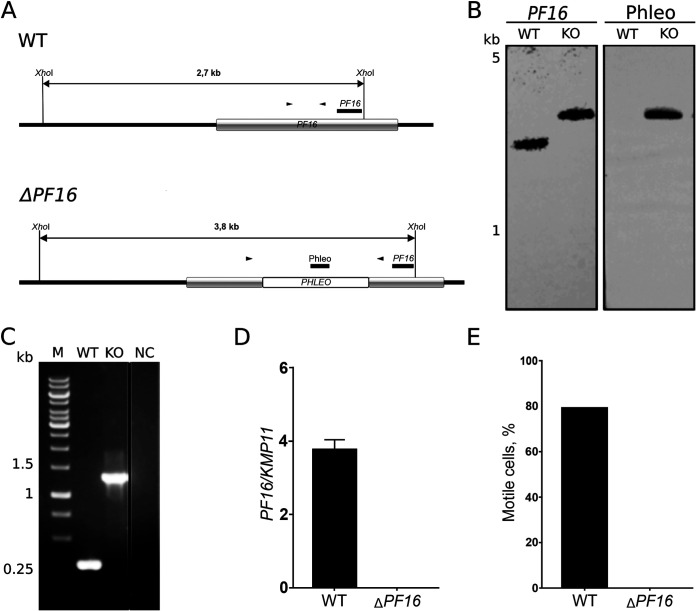
FIG 4.
Genetic ablation of PF16 using CRISPR-Cas9-mediated approach in N. esmeraldas. (A) Schematic representation of the wild-type (top) and recombined (bottom) PF16 alleles of N. esmeraldas. Expected sizes of DNA fragments after XhoI digestion and positions of the annealed probes and primers for diagnostic PCR are indicated. (B) Southern blot analysis of the XhoI-digested N. esmeraldas genomic DNA of the WT and PF16-ablated strains with PF16 and Phleo probes. (C) Diagnostic PCR for the wild-type and recombined (KO) PF16 alleles of N. esmeraldas. NC, negative control. (D) RT-qPCR control for the wild-type and ΔPF16 alleles of N. esmeraldas. (E) Percent motile cells in the wild type and ΔPF16 N. esmeraldas.