FIG 1.
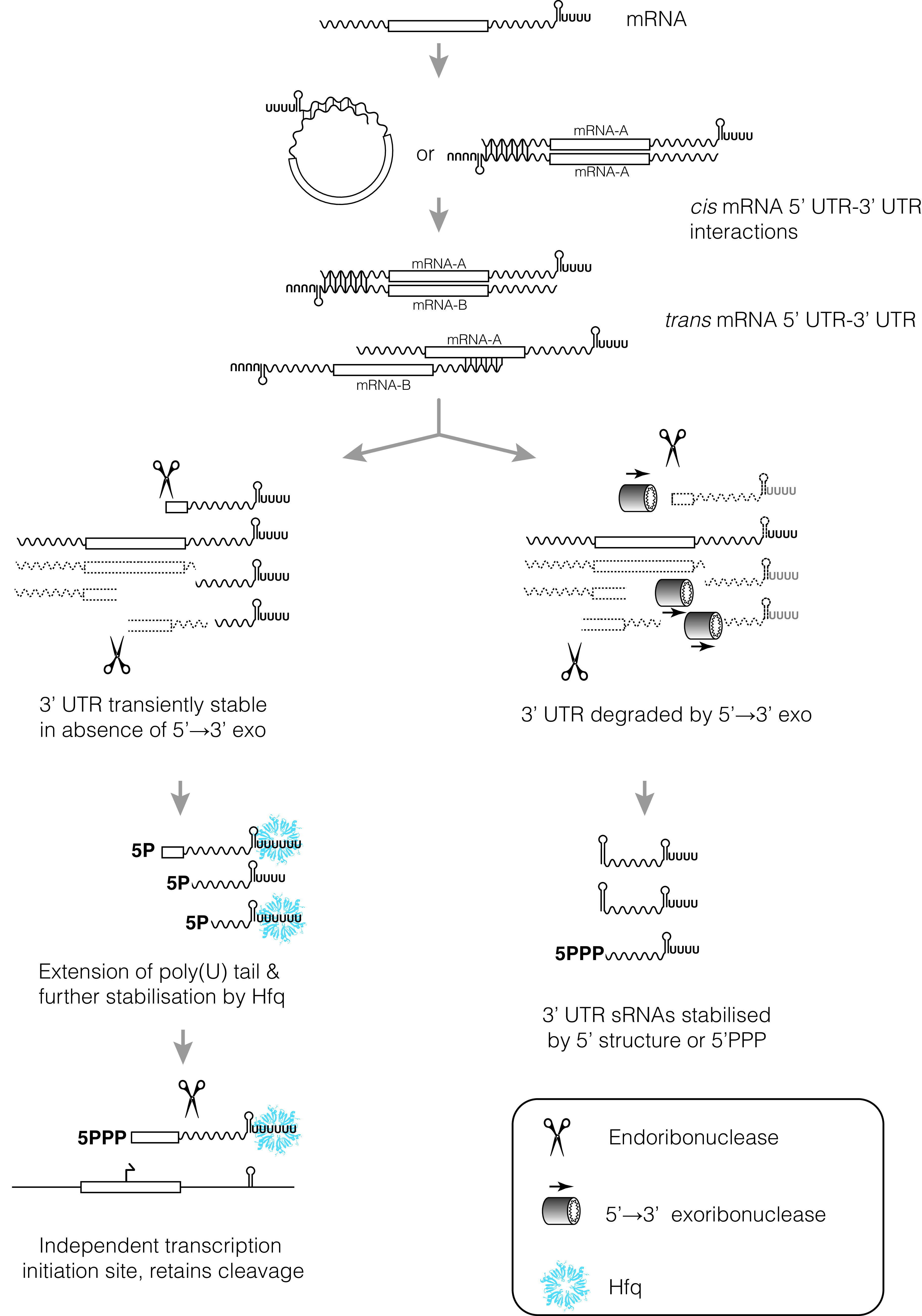
A continuum of regulatory 3′ UTR independence (see the text for a detailed description and examples). The UTRs of bacterial mRNAs are highly variable and contribute to expression of the coding sequence. Selected 3′ UTRs have been found to interact with cis-encoded 5′ UTRs, trans-encoded 5′ UTRs, and CDSs to control mRNA stability. The regulatory functions of many 3′ UTRs appear to have been physically separated from the mRNA. For bacteria that lack 5′→3′ exoribonuclease activity, cleaved 3′ UTRs will be stabilized by the intrinsic terminator alone (left branch). This may allow cleaved 3′ fragments to acquire additional stabilizing features, like an extended poly(U) tail that recruits the match-making sRNA chaperone, Hfq. As the regulatory 3′ UTR acquires more mRNA targets, additional regulatory inputs (like internal transcription start sites) may allow further separation of 3′ UTR and mRNA functions. (Right branch) In bacteria that encode 5′→3′ exoribonuclease activity, mRNA 3′ UTRs are efficiently degraded from the 5′ end. These 3′ UTRs must first acquire stabilizing 5′ structures (stems that sequester the 5′ end), or internal promoters that deposit protective 5′ triphosphates, before they are stabilized.
