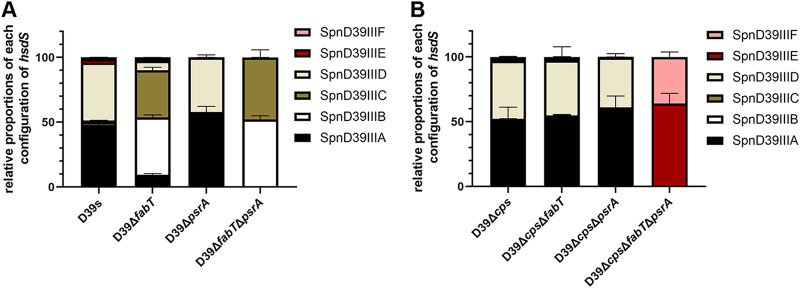
FIG 7.
Impact of PsrA and FabT on the hsdS allelic configuration. (A and B) Relative proportions of hsdS allelic genes in (A) encapsulated and (B) unencapsulated D39 derivatives were determined by qRT-PCR with cDNA as the templates. The transcripts of the 5′ noninvertible sequence of hsdS were detected in all strains as a reference to calculate the ΔCT values. ΔCT values of each hsdS allele were normalized again with the mean value of hsdSA as a reference in each strain to obtain ΔΔCT. The sum of the ΔΔCT of all six hsdS alleles was defined as 100%, and the relative composition of each hsdS allelic gene was calculated. Primers are listed in Fig. S6.