FIG 1.
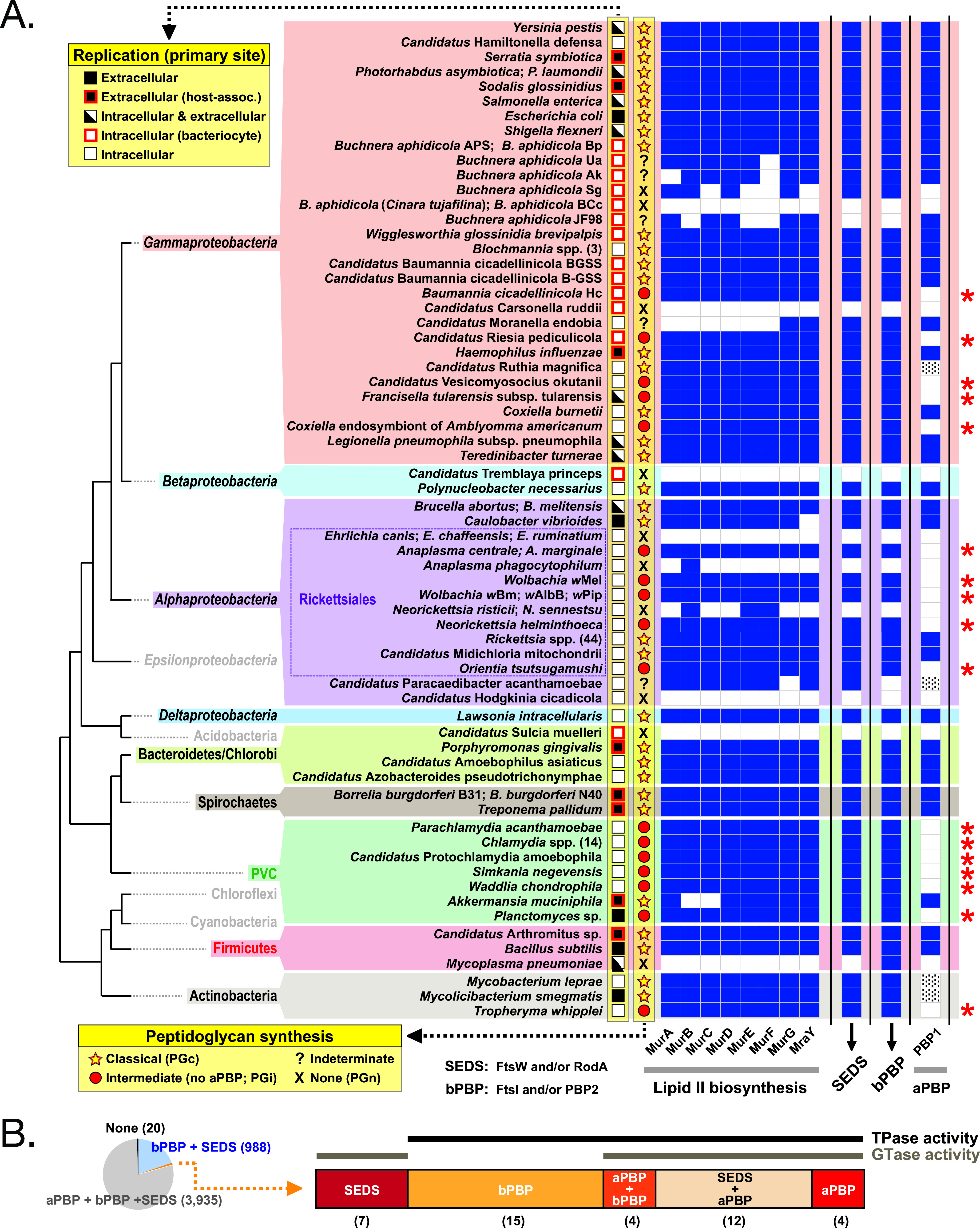
The class A PBP gene has been lost at least four times during evolution. (A) Analysis of key genes in the peptidoglycan biosynthesis pathway showing their presence or absence in obligate intracellular, facultative intracellular, host-associated, and free-living bacteria across 9 major bacterial groups. The predicted peptidoglycan (PG) status is shown as well as the primary site of replication of the bacteria. Genes involved in lipid II biosynthesis, SEDS family, class B PBPs, and class A PBPs were analyzed. The presence of a gene is shown by block color, the absence in white, and genes identified manually (not annotated as aPBP in KEGG) are shown with dashes. Bacterial organisms in the Gammaproteobacteria (Baumannia cicadellinicola Hc, “Candidatus Vesicomyosocius okutanii,” Francisella tularensis subsp. tularensis, Coxiella endosymbiont of Amblyomma americanum), Alphaproteobacteria (Anaplasma spp., Wolbachia spp., Orientia tsutsugamushi), PVC (Parachlamydia acanthamoebae, Chlamydia spp., “Candidatus Protochlamydia amoebophila,” Simkania negevensis, Waddlia chondrophila, Planctomyces sp.), and Actinobacteria (Tropheryma whipplei) all lack class A PBP homologs and are predicted to build intermediate PG-like structures (PGi) as highlighted by a red asterisk. PVC, Planctomycetes, Verrucomicrobia, and Chlamydiae; SEDS, shape, elongation, division, and sporulation; PBP, penicillin binding protein. Details of SEDS and bPBPs are shown in Fig. S1. (B) Relative quantification of bacterial strains containing different combinations of aPBP, bPBP, and SEDS proteins. This analysis was carried out on the 4,985 closed bacterial genomes available on KEGG in February 2020. Most bacteria (n = 4,923) either harbor all PG polymerization machines (80%) or qualify as PGi species (20%); a minority of species with other distributions (n = 42) are shown at the right as follows: SEDS only (n = 7), bPBP only (n = 15), bPBP and aPBP (n = 4), SEDS and aPBP (n = 12), and aPBP only (n = 4). Numbers of species in each category are shown in brackets. Bacteria binned to each category are listed in Table S1.
