FIG 2.
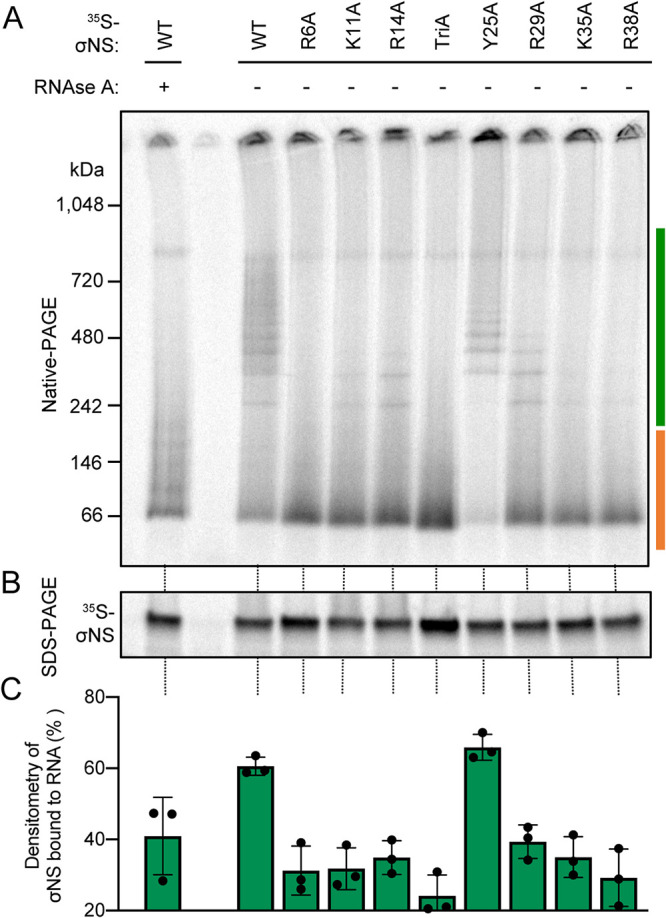
Alanine substitution of positively charged residues in a predicted RNA-binding domain of σNS alters RNA-dependent oligomerization. 35S-labeled σNS was expressed in rabbit reticulocyte lysates (RRLs) and incubated with or without RNase A. Samples were resolved by native PAGE to preserve oligomeric species (A) or SDS-PAGE to monitor protein expression (B) and visualized by autoradiography. The scale bar to the left of the native polyacrylamide gel (A) marks the kilodalton (kDa) ranges used for densitometric analysis of each σNS construct. The green scale bar marks σNS bound to RNA, whereas the orange scale bar marks unbound σNS. (C) The efficiency with which each σNS construct forms RNA-dependent oligomers was calculated by dividing the density of σNS bound to RNA (panel A, green scale bar) by the total density of σNS present in the gel (panel A, green and orange scale bar).
