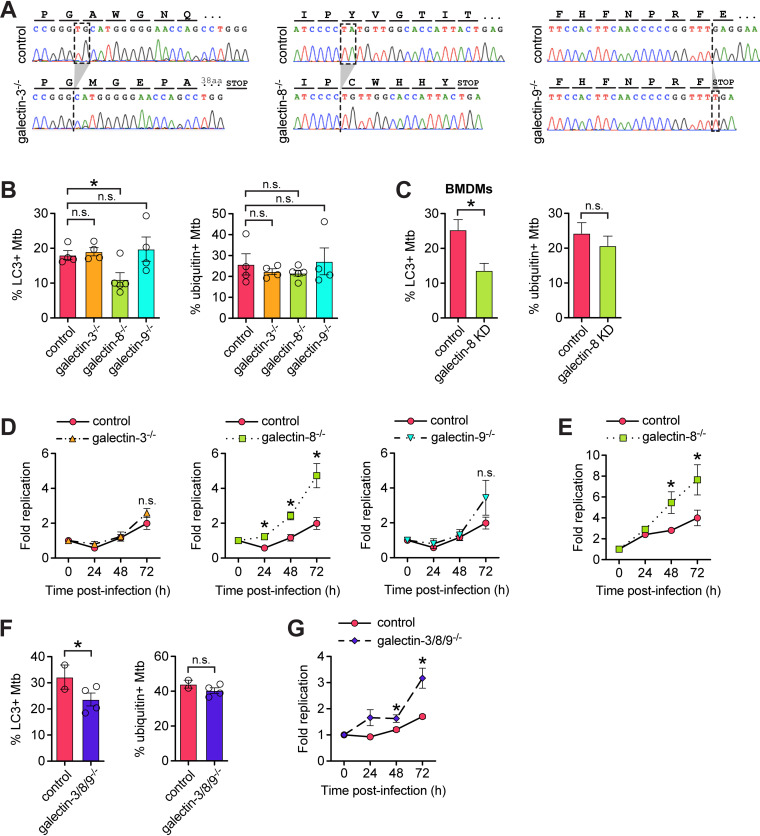
FIG 3.
Galectin-8 is required to efficiently target Mtb to selective autophagy to control Mtb replication in macrophages. (A) Representative chromatograms from galectin-3, -8, and -9 knockout cells lines indicating the nature of nonsense mutations introduced via CRISPR/Cas9. (B) Quantification of LC3- and ubiquitin-positive Mtb in control (sgRNAs targeting GFP) or individual galectin knockout RAW 264.7 cell lines 6 h postinfection. Circles represent data for each clonally selected cell line. (C) Same as in panel B but in BMDMs with galectin-8 knocked down via siRNA. (D) Fold replication of luxBCADE Mtb (MOI = 1) in control and knockout RAW 264.7 cells at the indicated time points. Data normalized to t = 0 h and representative of at least three independent experiments. (E) Same as in panel D but with replication measured by enumerating CFU. (F and G) Same as in panels B and D but with RAW 264.7 cell lines in which all three galectins are knocked out via CRISPR/Cas9. Error bars indicate the SEM of knockout cell lines or the SD for knockdown cells; for immunofluorescence (IF), at least 300 bacteria per cell line were assessed. *, P < 0.05; n.s., not significant.