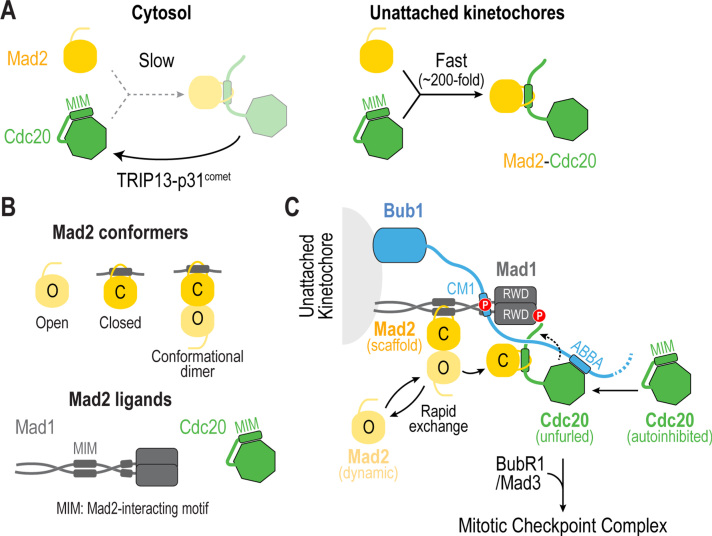
Fig. 4.
Catalysis of Mad2-Cdc20 complex formation at unattached kinetochores. (A) (left) Complex formation between Mad2 and Cdc20 is kinetically disfavored and subjected to disassembly by TRIP13-p31comet. (right) During mitosis, unattached kinetochores accelerate the rate of Mad2-Cdc20 complex formation by ~200 fold. (B) Mad2 has two different conformations, Open (O) and Closed (C), which interact with each other through dimerization. C-Mad2 is found bound to its ligands Mad1 and Mad2, which possess Mad2-interacting motifs (MIMs). (C) Proposed mechanism for how unattached kinetochores catalyze formation of the Mad2-Cdc20 complex. Mad2 is at two populations at the kinetochore: one that is stably bound to Mad1 (Mad2 scaffold) and a second that is cytosolic, transiently gets recruited to kinetochores through dimerization and becomes linked to Cdc20 (Mad2 dynamic). Kinetochores catalyze the transfer of Mad2 dynamic to Cdc20 by (1) recruiting Mad2 and Cdc20 to kinetochores, (2) positioning Cdc20 in close proximity to Mad2, and (3) unfurling Cdc20 to expose its MIM and promote the formation of the Mad2-Cdc20 complex.