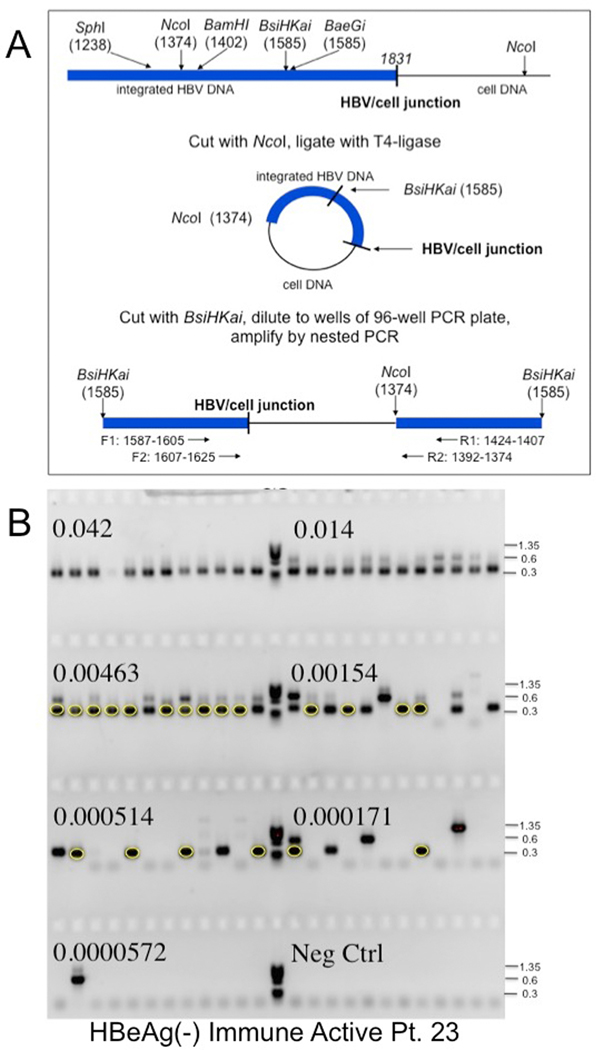
Figure 1: Inverse PCR detection of integrated HBV DNA.
A) Strategy for detection of integrated HBV DNA and clonal hepatocyte expansion. Inverse PCR, as used by Summers et al.,25, 28 was designed to detect the right hand junction of integrations of HBV dslDNA, the predominant precursor for integration, into host DNA.23, 24 Following cleavage and ligation (Figure 1A), the DNA samples were serially diluted and subjected to nested PCR using the indicated forward and reverse primers (Figure 1B). Primers are indicated in Supplementary Table 1 and Materials and Methods. (Figure 1A modified from reference by Mason et al.25). B) Gel electrophoresis of inverse PCR products. Samples from nested PCR, carried out in a 96 well tray, were subjected to gel electrophoresis in a 1.3% agarose gel. PhiX phage DNA digested with HaeIII was used as a size marker (M). The fraction of the initial DNA sample distributed across each row of 12 wells is indicated. Bands were picked from the last 5 rows, not including the negative control, and subjected to DNA sequencing to identify virus/cell DNA junctions. For instance, the circled bands arise from a single hepatocyte clone; other clones were also identified by DNA sequencing (not highlighted).