Figure 1 -.
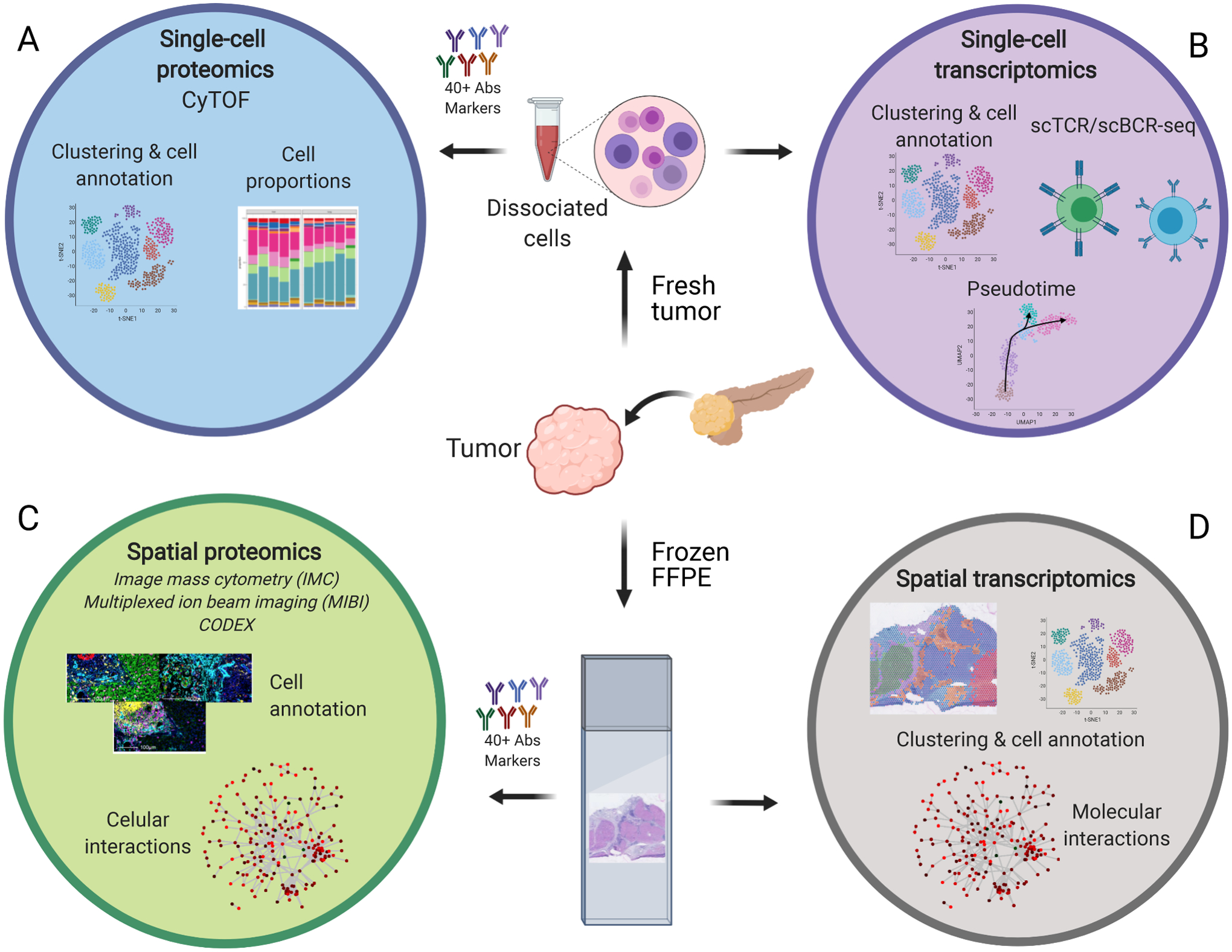
High dimensional transcriptomics and proteomics approaches for cancer profiling. Several high dimensional approaches are currently available to understand cancers cellular composition and inter-cellular interactions. A. Single-cell proteomics (CyTOF) provides cell composition and cell state information. B. Single-cell transcriptomics allows the same type of analysis, but its genome-wide coverage can also deliver cell trajectory predictions and T and B cell repertoires. In order to correlate cell composition and states to cellular interactions, spatial technologies are more informative than single-cell suspension analysis. C. With spatial proteomics and its single-cell resolution, it is possible to identify individual cell types and determine specific cell-to-cell interactions. D. Although it lacks single-cell resolution, spatial transcriptomics can predict cell interactions based on the molecular expression of receptors and ligands between different cell neighbors and discover driving oncogenic pathways among the different cell niches because it is not restricted to previously selected markers. The selection of which approach to apply will depend on what samples are available, how they are preserved, and what biological questions need answered.
