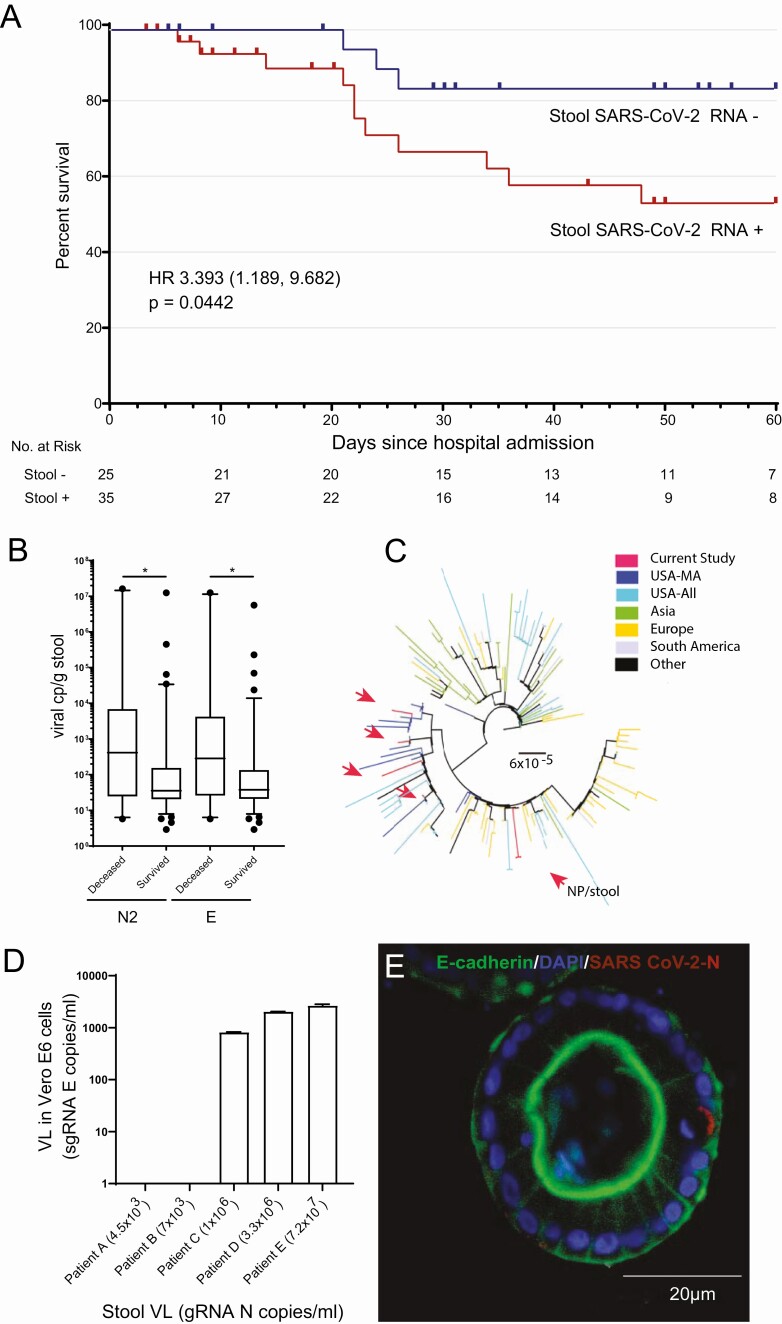
Figure 1.
Association of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the gastrointestinal (GI) tract with coronavirus disease 2019 (COVID-19) survival and tissue inflammation. A, Kaplan-Meier survival curve shows a significant increase in the mortality of COVID-19 patients with detectable stool SARS-CoV-2 RNA compared to those without detectable RNA. B, SARS-CoV-2 viral load was higher in those who died from COVID-19 than those who survived as measured by quantitative reverse-transcription polymerase chain reaction (qRT-PCR) of N2 and E gene RNA levels. *P <0.05. C, Phylogenetic tree showing stool-derived SARS-CoV-2 isolates (red arrows), along with representative sequences from Massachusetts (USA-MA), the United States (USA-All), Asia, Europe, South America, and Africa/Canada (Other). The scale represents 0.00006 nucleotide substitutions per site. D, SARS-CoV-2 isolated from COVID-19 patient stool replicated in Vero E6 cells. Subgenomic E RNA copies/mL was measured in Vero E6 cell lysates by qRT-PCR following inoculation with stool filtrates from COVID-19 patients with varying viral load. E, Immunofluorescence image of a representative colon organoid derived from surgically resected colon of a COVID-19 patient stained for DAPI (blue), E cadherin (green), and SARS-CoV-2 N (red). Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; gRNA, genomic RNA; HR, hazard ratio; NP, nasopharyngeal; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; sgRNA, subgenomic RNA; VL, viral load.