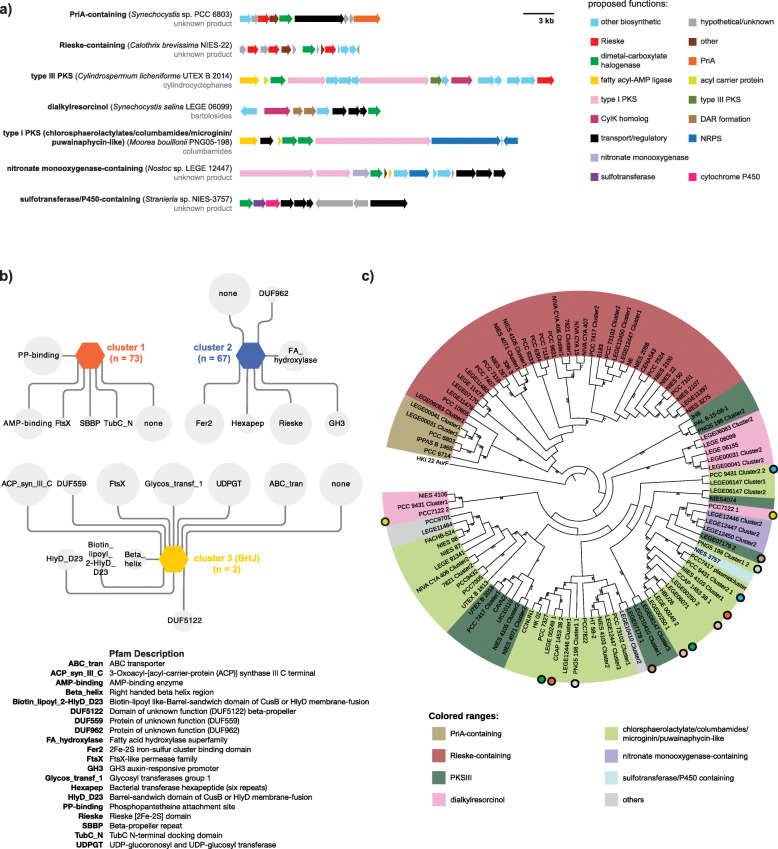
Fig. 4.
Diversity and genomic context of CylC-like enzymes BGCs. a Examples of the different BGC architectures found among the clusters encoding CylC homologs. b Genome Neighborhood Diagram (GND) depicting the Pfam domains associated with each cluster from the initial SSN of CylC homologs. The size of each node is proportional to the prevalence of the Pfam domain within the genomic context of the CylC homologs from each SSN cluster. c RAxML cladogram (1000 replicates, shown are bootstrap values > 70 %) of CylC homologs. The different colors represent a categorization based on common genes found within the associated biosynthetic gene clusters (see legend). Some BGCs contain more than one cylC homolog: circles of the same color depict different CylC homologs encoded in the same BGC. AurF (Streptomyces thioluteus HKI-22) was used as an outgroup