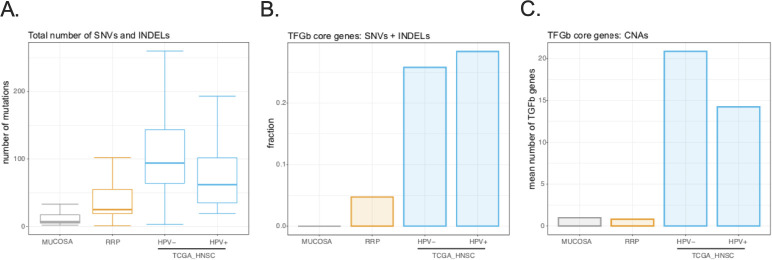
Figure 4.
Assessment of genomic alterations in papillomas and head and neck carcinomas. (A) Boxplot shows the total number of single nucleotide variants (SNVs) and small insertions and deletions (INDELs) called from whole exome sequencing using an ensemble approach was determined for normal laryngeal mucosa (n=8), papillomas (n=21), HPV-negative HNSCC (n=74) and HPV-positive HNSCC (n=314). (B) Barplot shows the fraction of samples from each cohort harboring an SNV or INDEL in one or more of the 43 TGF-b superfamily genes was determined. (C) Barplot shows the mean number of TGF-b superfamily genes amplified or deleted within each cohort was determined. HNSCC, head and neck squamous cell carcinoma; HPV, human papillomavirus; TGF-b, transforming growth factor-beta.