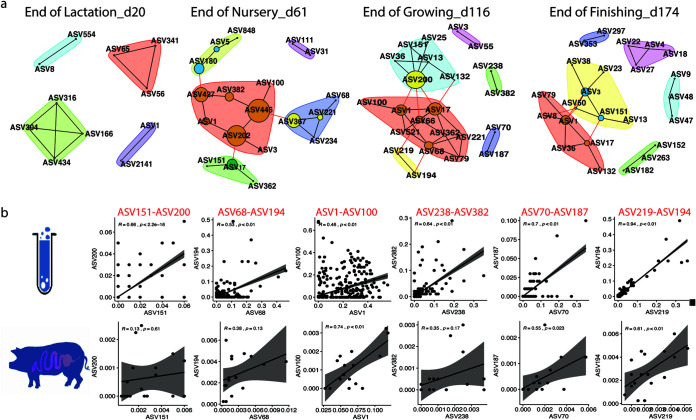
FIG 5.
Shared cooccurrence networks disclosed by SparCC (|R| > 0.4, P < 0.05) in both culture-independent (CI) and -dependent (CD) models at each time point (a). The SparCC algorithm within the Mothur software package was used to construct bacterial pairwise correlations in both the culture-enriched molecular profiling and live pig models. Correlations with coefficiency over 0.4 or less than −0.4 (P value less than 0.05) were included for downstream network display. Age-dependent ASVs with cooccurrence that were shared in both culture and live animals were clustered based on the Girvan-Newman algorithm. Sizes of nodes were determined by the betweenness-based centrality. Correlations of six pairs of cooccurring bacterial ASVs on d116 are shown in both CD (upper panel) and CI (lower panel) models (b). The six pairs of ASVs are ASV200 (Dorea)-ASV151 (Oribacterium), ASV68 (Bifidobacterium)-ASV194 (Lactobacillus), ASV1 (Megasphaera)-ASV100 (Acidaminococcus), ASV238 (Mitsuokella)-ASV382 (Mitsuokella), ASV70 (Lactobacillus)-ASV187 (Lactobacillus), and ASV219 (Lactobacillus)-ASV194 (Lactobacillus).