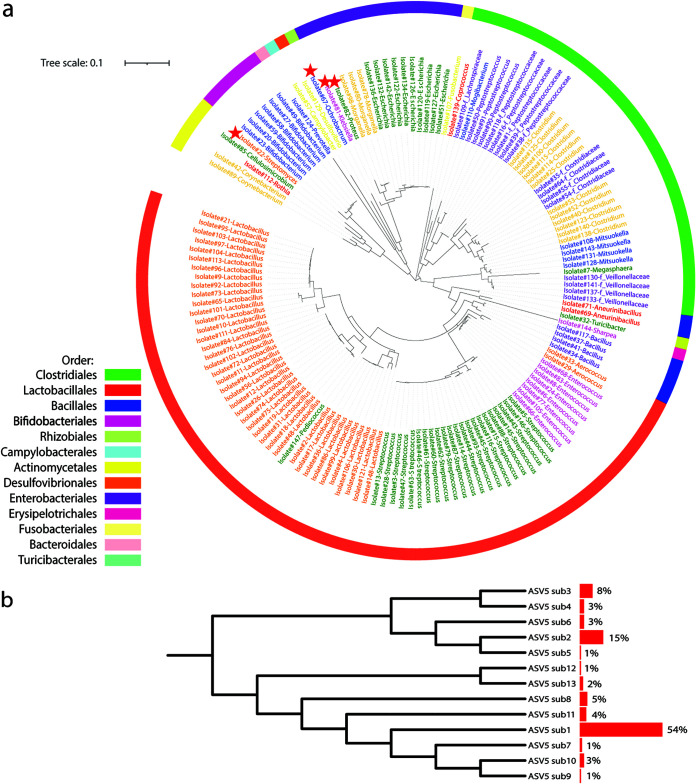
FIG 6.
(a) Phylogenetic tree analysis of 148 different bacterial taxa based on 16S rRNA gene V3-V7 hypervariable regions. Colonies were picked from eight agars (aerobic BHI2 and PEA and anaerobic B2I, BBE, BEEF, BSM, MGAM, and PEA). Isolates were identified by colony-PCR using a 16S rRNA gene primer set, 27f and 1492r. PCR amplicons were validated by gel electrophoresis and cleaned up for Sanger sequencing. Forward and reverse sequences were aligned to obtain near-full-length 16S rRNA gene contigs. All contigs were truncated to 16S rRNA gene V3-V7 (F, 5′-TACGGRAGGCAGCAG-3′, and R, 5′-GTAGCRCGTGTGTMGCCC-3′) before phylogenetic tree analysis using the Qiime2 program. The outermost ring was colored by orders. The clades of the tree were colored by genus. Node with a red star possesses a less than 95% similarity to the NCBI BLAST online database, indicating a potentially novel strain in the swine gut microbiome. (b) Phylogenetic tree of 13 sub-ASVs classified with ASV5 Lactobacillus. Side bar charts showing the contribution of each sub-ASV to the total ASV5 colony numbers.