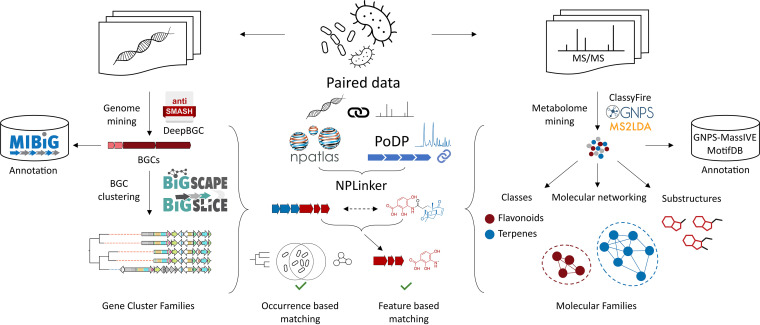
FIG 1.
Current state-of-the-art ecosystem of genomics (left) and metabolomics (right) natural product research, brought together by paired omics approaches (middle). Genomes are mined for biosynthetic gene clusters (BGCs) through tools such as antiSMASH and DeepBGC, and BGCs with structurally characterized products are stored in databases like MIBiG. BGCs are clustered into families with BiG-SCAPE and BiG-SLiCE. To infer compound classes, molecular families, and substructures, metabolomes (represented by collections of MS/MS spectra) are mined with tools such as ClassyFire, GNPS, MS2LDA, and MolNetEnhancer. Structural annotations relevant for microbiome research are stored in databases such as NP Atlas and MotifDB, and reference spectra are available in repositories such as GNPS-MassIVE. Paired data stored in platforms such as the Paired Omics Data Platform (PoDP) combine the two sides, which facilitates multi-omics approaches such as NPLinker that links gene cluster families (GCFs) to molecular families (MFs) through sample occurrence (also known as strain correlation) and feature-based matching.