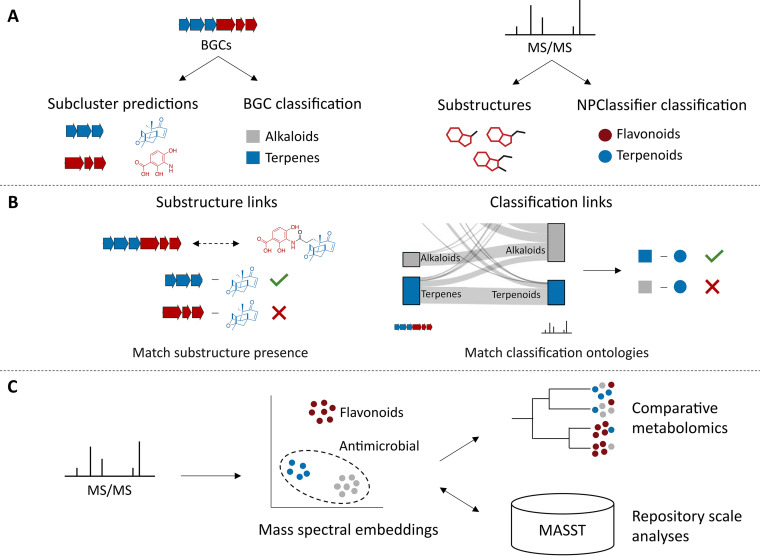
FIG 2.
Current and envisioned advances in multi-omics natural product discovery research. (A) Improved detection of subclusters and relevant natural product-related chemical compound classes in BGCs and MS/MS spectra will become possible based on machine learning-based computational tools. (B) We envision combining the existing BGC-metabolite matching approaches with substructure and chemical class predictions in platforms such as NPLinker. NPClassifier is a novel ML-based class predictor that considers both structural features and historical relationships between metabolites as defined by natural product researchers. (C) Mass spectral embeddings learned by Spec2Vec and trained with MS2DeepScore will enable fast and improved spectral similarity scoring. The bases for these mass spectral embeddings are the relationships between mass fragments and neutral losses based on their presence/absence in a large set of mass spectra. We expect that these embeddings will allow the rapid annotation of classes, substructures, or other labels such as pathways or functions based on clustering techniques. Finally, the developed workflows can also form the basis for improved comparative and repository-wide metabolomics approaches that highlight shared and novel chemistry produced by microbiomes.