FIG 2.
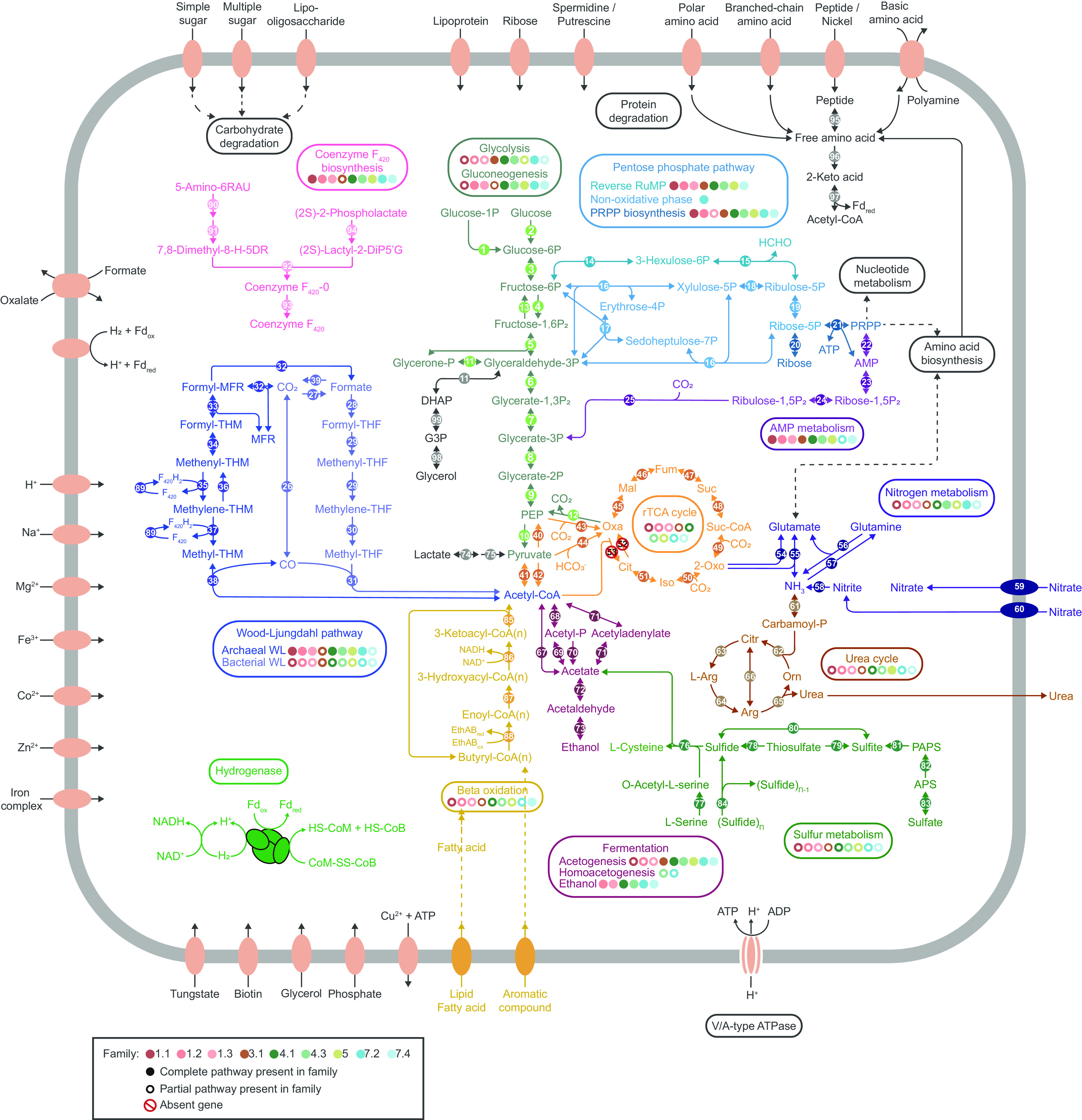
Overview of metabolic potentials in Ca. Bathyarchaeia from hot springs. Genes related to glycolysis, gluconeogenesis, the pentose phosphate pathway, AMP metabolism, the Wood-Ljungdahl pathway, the rTCA cycle, nitrogen and sulfur metabolism, the urea cycle, beta-oxidation of fatty acids, fermentation, coenzyme F420 biosynthesis, protein degradation, and membrane transporters are shown. Detailed gene copy information associated with above-mentioned pathways is in Data Set S2. Reverse RuMP, reverse ribulose monophosphate pathway; PEP, phosphoenolpyruvate; G3P, glycerol-3-phosphate; DHAP, dihydroxyacetone phosphate; PRPP, phosphoribosyl pyrophosphate; MFR, methanofuran; PAPS, 3′-phosphoadenylyl sulfate; APS, adenylyl sulfate; Fd, ferredoxin.
