FIG 1.
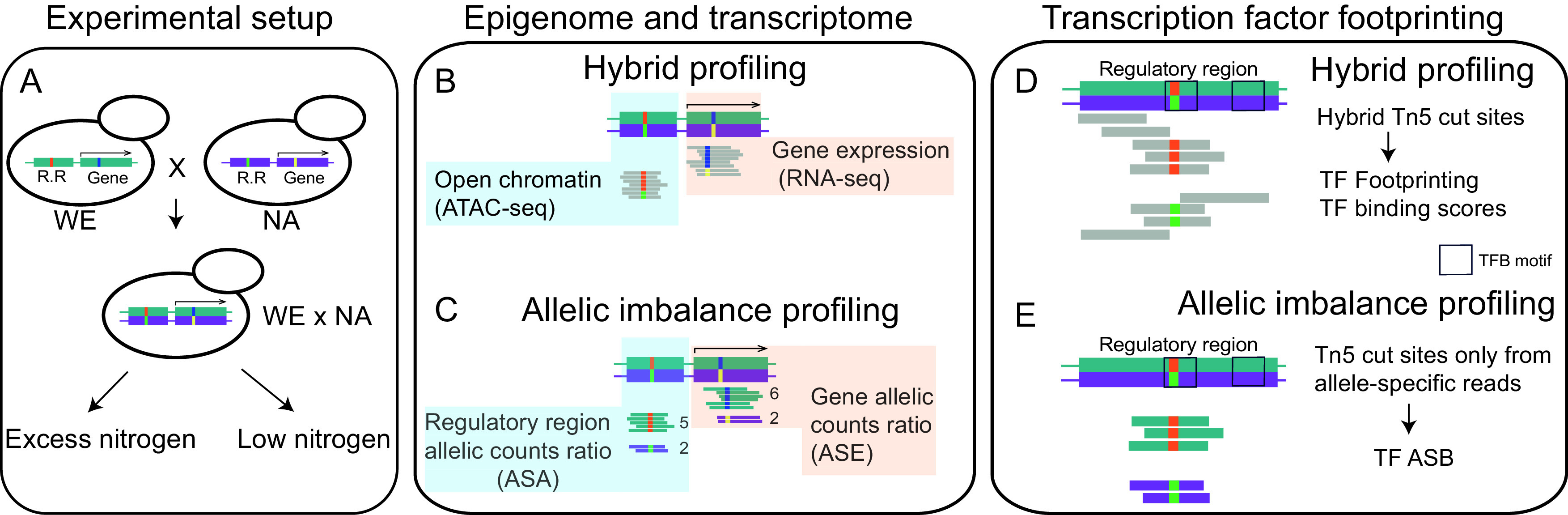
Allele-specific expression and chromatin accessibility profiling to reveal molecular mechanisms orchestrating gene expression divergence in response to nitrogen scarcity. (A) Representative haploid strains of the WE (Wine European) and NA (North American) lineages of S. cerevisiae were selected to construct a WE × NA hybrid which was used to perform fermentations in synthetic wine media under low and excess nitrogen conditions (SM60 and SM300, respectively). Large boxes denote a cis-regulatory region (R.R) and an open reading frame (ORF) (arrow on top), while smaller boxes depict polymorphisms occurring in each genetic background. (B) RNA-seq and ATAC-seq were employed to profile the hybrid's transcriptome and open chromatin landscape at regulatory regions between nitrogen conditions. Gray bars denote the common pool of hybrid reads. (C) RNA-seq and ATACseq reads that mapped to parent-specific single-nucleotide polymorphisms (SNPs) were used to determine allele-specific expression (ASE) and allele-specific accessibility (ASE and ASA, respectively). Reads highlighted in colors indicate different pools according to their parental origin. (D) ATAC-seq cut sites were used to infer in silico footprinting of transcription factor binding (TFB) in the hybrid. (E) Allele-specific ATAC-seq alignments were separated by parental origin to obtain allele-specific ATAC-seq cut sites which were used to infer allele-specific binding (ASB) from TFB in silico footprinting.
