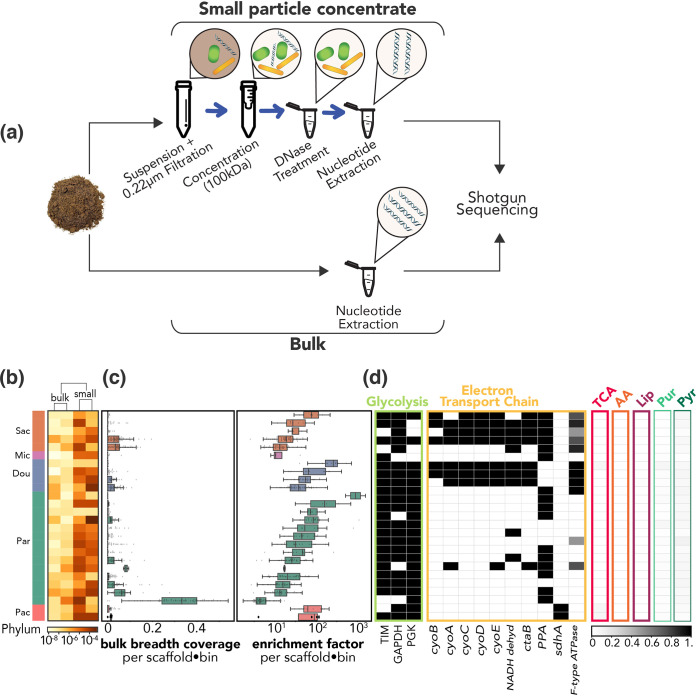
FIG 1.
Enrichment and metabolic profiles of CPR bacteria in soil concentrate metagenomes. (a) Method for concentration of small particles from soil for metagenomic sequencing (top) compared to sample preparation methods for bulk soil metagenomes (bottom). (b) Heat map showing relative abundance of 26 organisms by phylum across bulk metagenomes and concentrate metagenomes. Sac, Saccharibacteria; Mic, Microgenomates; Dou, Doudnabacteria; Par, Parcubacteria; Pac, Pacearchaeota. (c) Coverage-based metrics showing recovery and enrichment in all concentrates combined relative to that in bulk fractions, combined as boxplots. (Left) Breadth of coverage of scaffolds comprising each genome (bin) in the bulk fraction. (Right) Enrichment factor (i.e., relative abundance of a scaffold from the concentrate metagenome over a scaffold’s bulk metagenome relative abundance) for each genome. (d) Metabolic analysis of each genome, including (i) presence of each of three glycolysis genes that are highly conserved among CPR bacteria, (ii) genes involved in the electron transport chain (NADH dehydrogenase; ctaB, heme O synthase [EC 2.5.1.141]; PPA, inorganic pyrophosphatase [EC 3.6.1.1]; sdhA, succinate dehydrogenase/fumarate reductase, flavoprotein subunit [EC 1.3.5.1 1.3.5.4]), and (iii) percentage completeness (grayscale) of F-type ATPase, the TCA cycle (tricarboxylic acid cycle), and pathways for amino acid biosynthesis (AA), lipid biosynthesis (Lip), purine biosynthesis (Pur), and pyrimidine biosynthesis (Pyr).