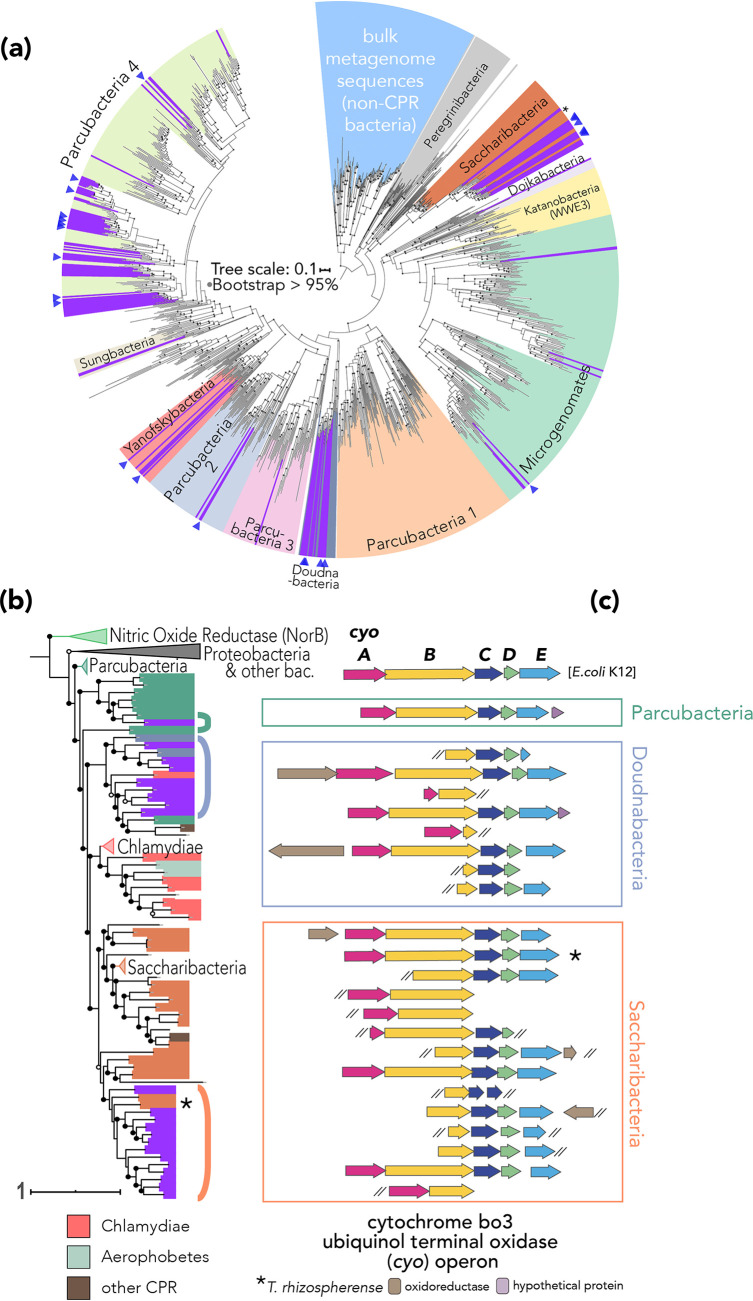
FIG 2.
Soil CPR phylogeny and cytochrome operon synteny. Sequences assembled from the small concentrate metagenomes are shown in purple. (a) RpS3 tree of CPR bacteria rooted using RpS3 sequences that were assembled from bulk metagenomes, in light blue. Blue triangles denote draft genome recovered. Nodes with bootstrap values greater than or equal to 0.95 are marked as filled black circles. (b) Phylogenetic relationships of cytochrome bo3 ubiquinol terminal oxidase subunit I across bacterial phyla. Circles overlaid on nodes correspond to support values (unfilled, >0.50; filled, >0.70). *, the placement of T. rhizospherense CyoB (b) and its operon in (c) (6). Brackets next to tree tips correspond to phyla by color (green, Parcubacteria; blue, Doudnabacteria; orange, Saccharibacteria) and to sequence order in the synteny diagram (c). Tree rooted using a heme-copper oxidase superfamily member, the nitric oxide reductase (NorB). (c) Synteny diagram of cytochrome ubiquinol oxidase operon genes (cyoA, cyoB, cyoC, cyoD, and cyoE) with operon from E. coli K-12 as a reference. Scale bars correspond to the average number of substitutions per site across alignment.