FIG 2.
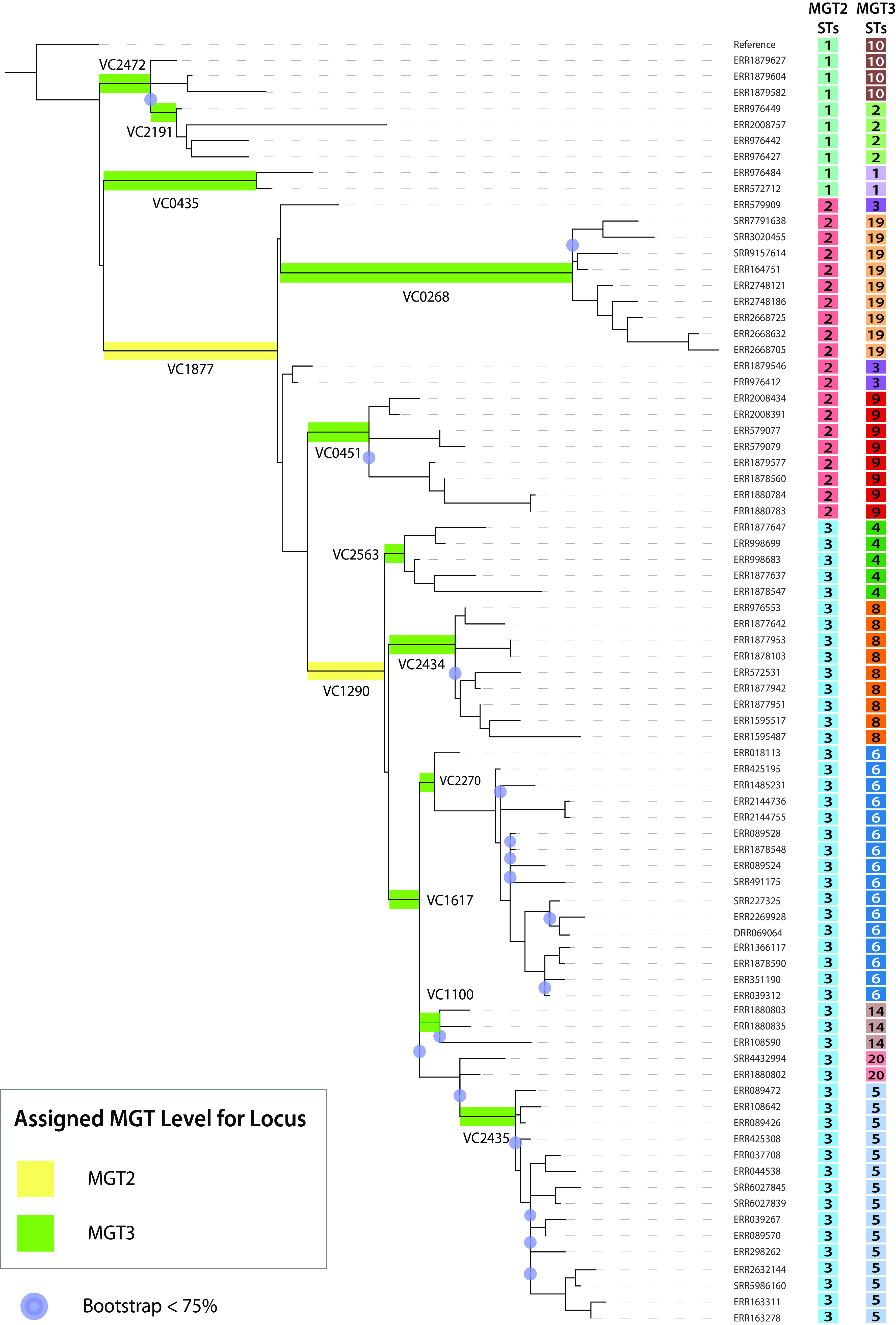
Selected MGT2 to MGT3 loci overlayed onto representative seventh pandemic phylogeny. A maximum-likelihood phylogeny was created from 80 representative isolates selected across the seventh pandemic (7P) using RAxML (42). Branches are highlighted with species core genes separating 7P clades. Mutations in genes within MGT2 and MGT3 are highlighted in yellow and green, respectively. The STs defined at MGT2 and MGT3 levels are aligned next to each isolate. Genetic relationships were derived from single-nucleotide polymorphism (SNP) differences in the 3,759 core 7P pandemic loci. The tree was rooted on the V. cholerae N16916 reference genome. Branches were bootstrapped 1,000 times, and branches showing less than 75% support are marked with a purple circle.
