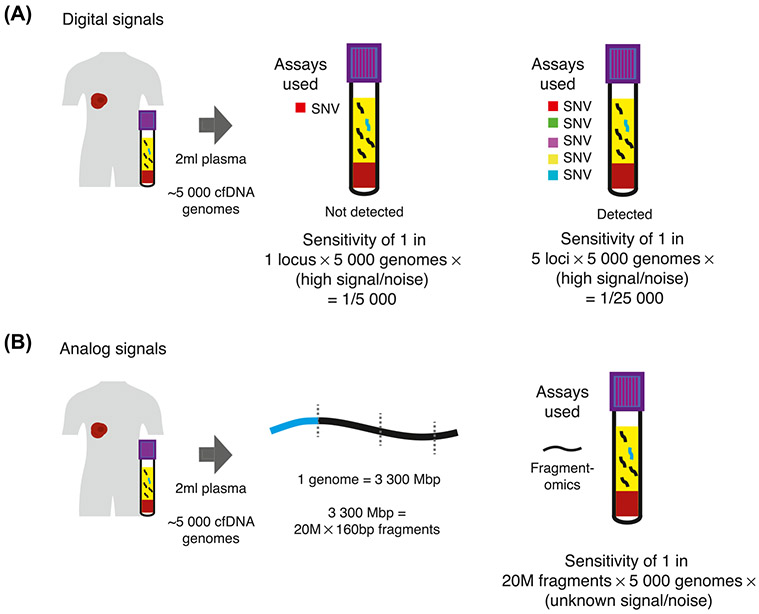
Figure 2. Determinants of Sensitivity for Digital and Analog Cancer Signals.
(A) For digital cancer signals, such as SNVs, the sensitivity achieved approximates the number of loci targeted multiplied by the number of genome equivalents targeted. The signal-to-noise ratio of such digital signals is high, up to 1:10K molecules for digital PCR assays, and potentially one to two orders of magnitude further for error-suppressed sequencing [4,26]. (B) For analog signals such as fragmentation patterns, there are 20 × 106 cfDNA fragments (with a median fragment size of 166 bp) for each genome equivalent. Therefore, the total number of reporters that may be targeted is maximized, though the signal-to-noise ratio is far lower than a digital signal. Due to the large numbers of reporters, even moderate signal-to-noise ratios may provide high sensitivity for cancer. Abbreviations: cfDNA, cell-free DNA; SNV, single nucleotide variant.