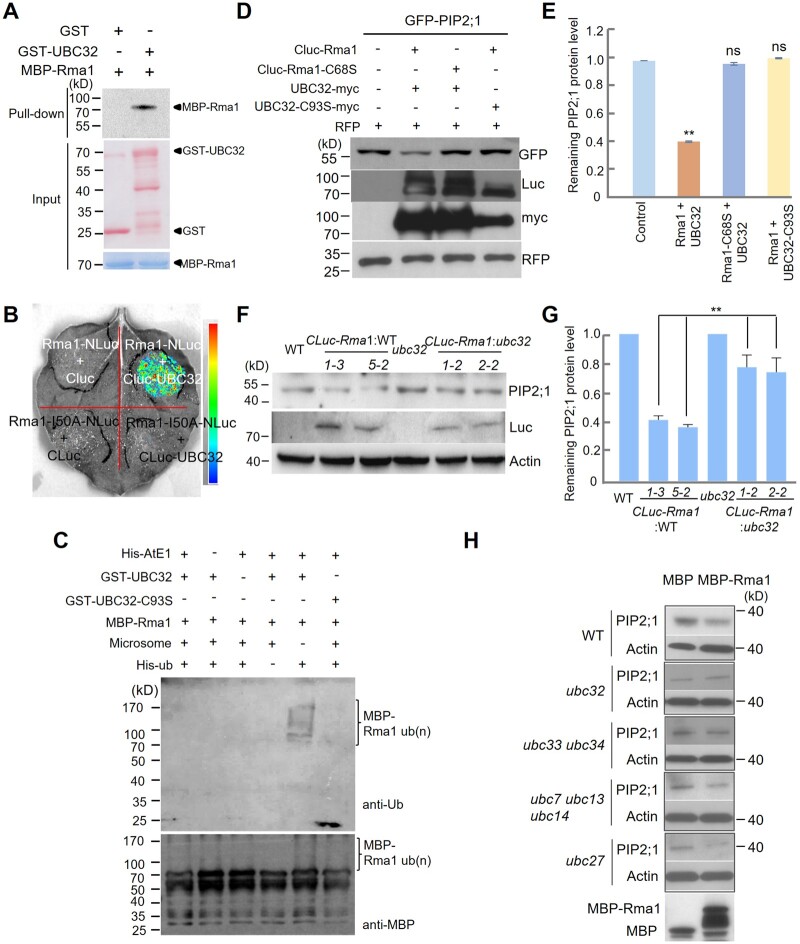
Figure 4.
UBC32 and Rma1 function as an E2–E3 pair to ubiquitinate and degrade PIP2;1/PIP2;2. A, Rma1 interacts with UBC32 based on a pull-down assay. GST, GST-UBC32, and MBP-Rma1 expressed in E. coli were used in this pull-down assay. Equal amounts of GST and GST-UBC32 bound to anti-GST resins were incubated with 2-μg MBP-Rma1. Pull-down products were detected using anti-MBP antibody. The arrowheads on the right indicate the positions of full-length GST/MBP-tagged proteins. B, Rma1 binds to UBC32 in planta based on an LCI experiment. CLuc-UBC32 interacts with Rma1-NLuc, but not the mutated RING domain form (Rma1-I50A-NLuc). The pseudocolor bar on the right shows the range of luminescence intensity. C, In vitro autoubiquitination assay of Rma1. MBP-Rma1, His-AtE1, Ub, and 200 ng microsome were combined with GST-32 or GST-UBC32-C93S, respectively. The reaction was carried at 30°C for 90 min, and immunoblot analysis was performed using anti-MBP and anti-ub antibodies. D, The E3 ligase activity of Rma1 and E2 activity of UBC32 are necessary for the degradation of PIP2;1. Single amino acid mutations Rma1-C68S and UBC32-C93S, which lost E3 ligase activity or E2 activity, respectively, were used in this assay. Agrobacterium containing the GFP-PIP2;1 plasmid was co-injected into N. benthamiana with Agrobacterium containing the corresponding plasmids as indicated. Total protein was extracted from N. benthamiana leaves at 3 days post infiltration and analyzed by immunoblot analysis. RFP was used as the co-expressed control in this assay. E, Quantification of GFP-PIP2;1 protein levels in (D). Values represent the mean ± sd (n = 3, n means biological replicate). Statistical significance was determined by one-way ANOVA (**P <0.01; ns, no significant difference). F, The regulation of PIP2;1 by UBC32 is Rma1-dependent. PIP2;1 protein level was detected in WT, CLuc-Rma1:WT transgenic plants, ubc32 and CLuc-Rma1:ubc32 transgenic plants using anti-PIP2;1 antibody. G, Quantification of PIP2;1 protein levels in (F). Values represent the mean ± sd. Statistical significance was determined by one-way ANOVA (**P <0.01). H, PIP2;1 stability is not affected by other E2s in the ERAD system. Crude extracts of WT and different E2 mutants were incubated with MBP and MBP-Rma1 at 22°C and PIP2;1 protein was checked using anti-PIP2;1 antibody.