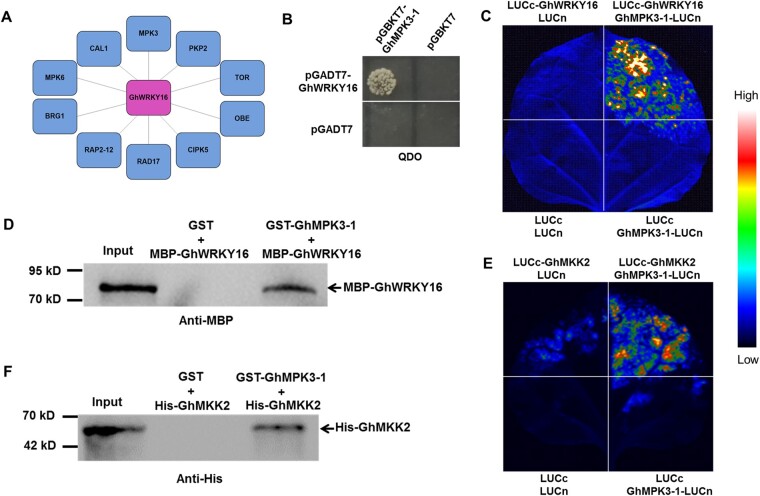
Figure 5.
Assay of interaction between GhWRKY16 and GhMPK3-1, and GhMKK2 and GhMPK3-1 in vitro and in vivo. A, Co-expression network of GhWRKY16, as determined from ccNET (http://structuralbiology.cau.edu.cn/gossypium/). B, Yeast two-hybrid assay of the interaction between GhWRKY16 and GhMPK3-1. Yeast transformants containing pGADT7-GhWRKY16 or pGBKT7-GhMPK3-1 were spotted onto SD–Leu–Trp after yeast mating, and interaction was tested on SD–Leu–Trp–His–Ade, using pGBKT7 and pGADT7 empty vectors as negative controls. C, LCI assay of the interaction between GhWRKY16 and GhMPK3-1. Constructs expressing GhMPK3-1-LUCn and LUCc-GhWRKY16 were co-infiltrated in N. benthamiana leaves, using LUCn and LUCc as negative controls. D, Pull-down assay of GhWRKY16 and GhMPK3-1. Recombinant MBP-GhWRKY16 was incubated with GST-GhMPK3-1 in vitro, using GST as negative control. Pulled-down proteins were analyzed by immunoblotting with anti-MBP antibody. E, LCI assay of the interaction between GhMPK3-1 and GhMKK2. Constructs expressing GhMPK3-1-LUCn and LUCc-GhMKK2 were co-infiltrated in N. benthamiana leaves, using LUCn and LUCc as negative controls. F, Pull-down assay of GhMPK3-1 and GhMKK2. Recombinant GST-GhMPK3-1 was incubated with His-GhMKK2 in vitro, using GST as negative control. Pulled-down proteins were analyzed by immunoblotting with anti-His antibody