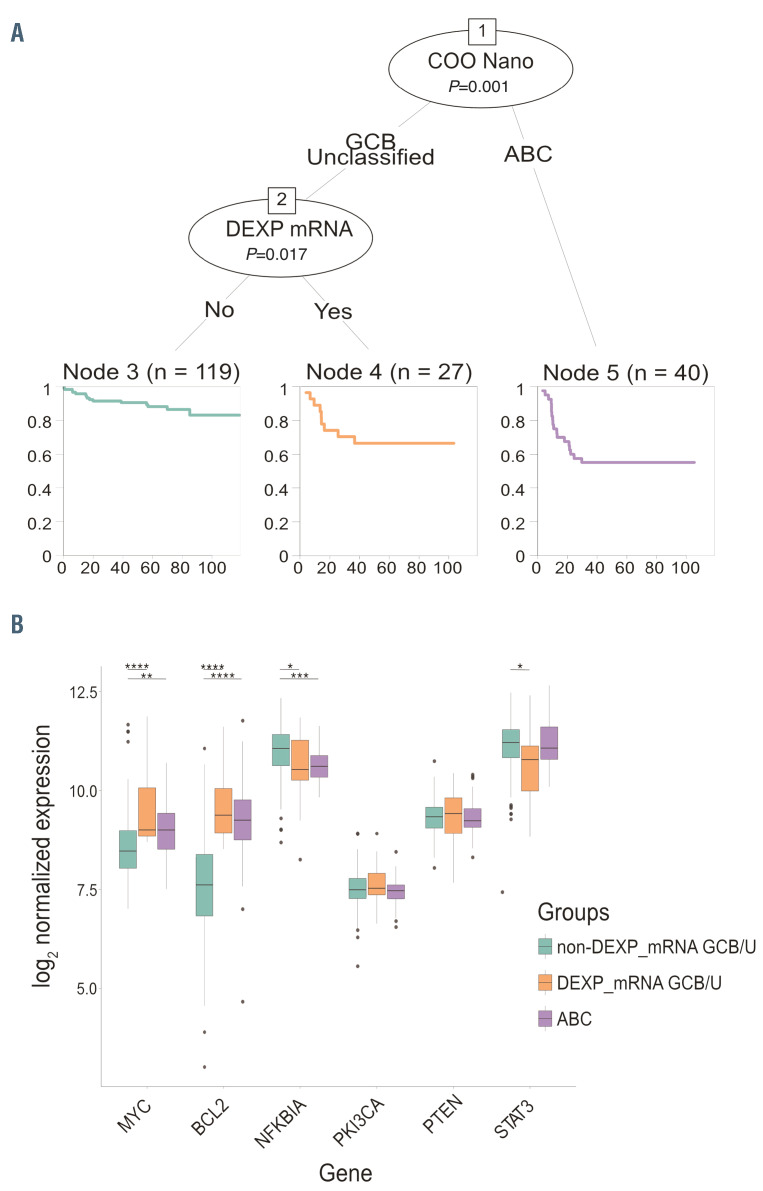
Figure 2.
Integrating cell of origin with MYC/BCL-2 DEXP_mRNA status for prognostication in diffuse large Bcell lymphoma. (A) Recursive partitioning analysis integrating cell of origin (COO) classification and DEXP_mRNA status, allowing segregation of patients in three main prognostic subgroups (a low risk non-DEXP-mRNA GCB/U subset, and two high risk groups: MYC/BCL-2-DEXP-mRNA GCB/U and ABC). (B) Box plot graphs indicating the expression levels of the additional targets included in the panel (MYC, BCL-2, NFKBIA, STAT3, PIK3CA, PTEN) in the three main patients subgroups identified by the recursive partitioning analysis (non-DEXPmRNA GCB/U, MYC/BCL-2 DEXP_mRNA GCB/U, ABCderived diffuse large B-cell lymphoma [DLBCL]. P-value was calculated with Student ttest by comparing non DEXP_mRNA GCB/U group, selected as a reference, versus other groups. ABC: activated B-cells; GCB: germinal center B cells; GCB/U: GCB unclassified; DEXP_mRNA: double expressor GCB/U DLBCL. COO Nano: COO as determined by T-GEP with NanoString profiling