FIGURE 1.
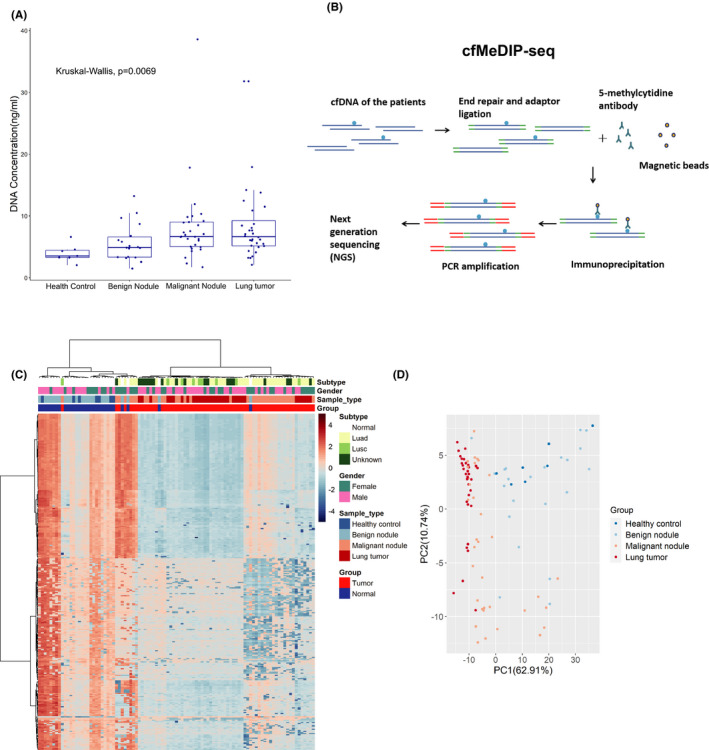
Cell‐free methylated DNA immunoprecipitation and high‐throughput sequencing (cfMeDIP‐seq) identified the differentially methylated regions (DMR) between lung tumors and normal controls. A, Cell‐free DNA (cfDNA) concentrations of patients from different groups. Box plots are displayed with a median center line; box range from the 25th to 75th percentile. B, Workflow of the cfMeDIP‐seq. cfDNA is ligated with sequencing adaptors. Methylated cfDNA fragments are immunoprecipitated with the 5‐methylcytidine antibody, followed by PCR amplification and next‐generation sequencing (NGS). C, Heatmap of the top 300 DMR identified in the plasma cfDNA between 30 normal controls and 67 lung tumor patients. Luad, lung adenocarcinoma; Lusc, lung squamous cell carcinoma. D, The top 300 DMR were used to generate the principal component (PC) plot to classify lung tumors and normal controls
