Abstract
Temozolomide (TMZ) resistance is the main challenge in the management of glioma patients. Heparanase can mediate the secretion and function of exosomes, which are considered to be a promising molecular delivery system for cancer therapy. Therefore, this study aimed to investigate whether heparanase‐mediated delivery of exosomes was related to TMZ resistance of glioma. Heparanase was upregulated in TMZ‐resistant glioma cells, and overexpression of heparanase led to increased resistance of U87 cells to TMZ. Knockdown of heparanase led to increased sensitivity of TMZ‐resistant U251 cells (U251R) cells to TMZ. Heparanase promoted the secretion of exosomes from glioma cells, and coculture with exosomes derived from heparanase knockdown U251R cells partly restored the sensitivity of U251 cells to TMZ compared with exosomes derived from si‐control transfected U251R cells. It was identified by circular RNA microarrays that hsa_circ_0042003 was upregulated in exosomes derived from U251R, which could be positively regulated by heparanase. U251R cell‐derived exosomal hsa_circ_0042003 conferred the resistance of U251 cells to TMZ. In vivo studies also showed that U251R cell‐derived exosomes induced resistance of U251 cells to TMZ, and the combination of tail‐injected exosomal si‐heparanase or exosomal si‐hsa_circ_0042003 and intraperitoneal TMZ applied to nude mice abolished TMZ resistance. Heparanase mediated the transfer of exosomal hsa_circ_0042003 from TMZ‐resistant glioma cells to drug‐sensitive cells, which contributed to the chemoresistance of glioma to TMZ.
Keywords: heparanase, glioma, chemoresistance, exosome, circRNA
The present study showed that heparanase contributed to the resistance of glioma to temozolide (TMZ), which involved its promotion of the secretion of TMZ‐resistant cell‐derived exosomes. Additionally, the si‐hsa_circ_0042003 delivered by exosomes derived from U251 cells could reverse the resistance of TMZ‐resistant glioma to TMZ, inhibiting cell proliferation and accelerating cell apoptosis, and restraining tumor growth in vivo. This study could provide ideas for the development of treatment strategies for TMZ‐resistant glioma patients.
1. INTRODUCTION
Glioma, as the most common primary malignant brain tumor in adults, has a high morbidity and mortality.1 Temozolomide (TMZ) is the first‐line treatment for glioma, but the development of chemoresistance limits its clinical efficacy.2, 3 Resistance to TMZ and subsequent tumor recurrence has become a major obstacle in the clinical therapy of glioma.4 High levels of O6‐methylguanine methyltransferase and DNA damage repair barriers could be part of the cause of TMZ resistance.4 However, the underlying mechanism of TMZ resistance in glioma remains unclear.
Heparanase is an endoglucuronidase, which can change the structure and function of heparan sulfate (HS) proteoglycans by cutting HS chains.5, 6 Studies have found that heparanase contributes to tumor‐mediated cell surface and ECM remodeling, and is closely related to tumor metastasis and angiogenesis.7, 8 Recent studies have confirmed that the expression of heparanase in a variety of cancers is upregulated, which is associated with increased metastasis and poor prognosis.8, 9, 10 High levels of heparanase are related to the poor prognosis of patients with high‐grade glioma; therefore, changing heparanase levels to target malignant brain tumor cells and their microenvironment could provide ideas for the treatment of malignant glioma.11 Increasing evidence also confirms that heparanase is involved in all processes of tumor formation, including chemoresistance.12 However, the effect of heparanase on glioma chemotherapy resistance is less reported.
Exosomes are nanoscale extracellular vesicles that contain a specific amount of RNA molecules, proteins, and lipids, which can be released by all cells and naturally exist in the blood.13 At present, exosomes are considered as the carriers of signal transduction between cells. Exosomes can be internalized by neighboring target cells and mediate cell communication by releasing their contents into the cellular microenvironment of the recipient cell.14 Increased data indicate that tumor‐derived exosomes are involved in the regulation of pathophysiological processes, including tumor growth, metastasis, microenvironment regulation, and chemoresistance.15, 16 Recent studies have found that circular RNAs (circRNAs) can be metastasized by exosomes, and exosomal circRNAs have the potential to become biological indicators for cancer diagnosis, treatment, and prognosis.17 A previous study reported that heparanase affects the composition of protein and mRNA in exosomes.18 However, it is unclear whether heparanase is involved in exosomal circRNAs in the process of mediating exosome transfer.
In this study, we aimed to explore whether heparanase‐mediated exosomal delivery is related to the TMZ resistance of glioma. We identified circRNAs related to TMZ resistance in glioma cell‐derived exosomes. Moreover, through a nude mouse xenograft model, we initially assessed whether drug‐resistant cells affect the resistance of sensitive cells to TMZ via exosomes in vivo. This study may provide new ideas for the development of potential therapies for TMZ‐resistant glioma.
2. MATERIALS AND METHODS
2.1. Serum samples
A total of 40 patients with advanced glioma from The Second Affiliated Hospital of Zhengzhou University were enrolled in the present study. Of them, 20 patients had progressive disease during primary chemotherapy or suffered recurrence within 6 months of completing primary chemotherapy (TMZ‐resistant patients), and 20 patients had recurrence beyond 6 months or were without relapse after TMZ‐based chemotherapy (TMZ‐sensitive patients). Assessment of each patient’s response to TMZ was based on RECIST version 1.1. Peripheral blood samples were collected from glioma patients prior to starting chemotherapy, and serum samples were obtained after centrifugation. There was no healthy donor serum used as control for the following serum exosome analysis. The study was approved by the Ethics Committee of The Second Affiliated Hospital of Zhengzhou University, and written consent was obtained from all patients.
2.2. Cell culture and treatment
The human malignant glioma cell lines (T98G, U138, LN382, KMG4, LN235, LN308, A172, AM38, U87, U251) were purchased from Chinese Academy of Sciences Cell Bank (Shanghai, China) and cultured in DMEM (Invitrogen Life Technologies) containing 10% FBS (Gibco), 100 U/mL penicillin, and 100 mg/mL streptomycin (Sigma‐Aldrich) at 37℃ with 5% CO2.
As described previously, the TMZ‐resistant glioma cell line (U251R) was obtained by gradually exposing the parental cell line (U251) to medium with increasing doses of TMZ.19 The medium containing TMZ was changed every 2‐3 days. In addition, 30 μM TMZ was used for subsequent analysis.
Small interference RNA targeting heparanase (si‐heparanase) and matching control, as well as si‐hsa_circ_0042003 and corresponding control, were obtained from GenePharma. Heparanase pcDNA3.1 plasmid (pcDNA‐heparanase), hsa_circ_0042003 pcDNA3.1 plasmid (pcDNA‐hsa_circ_0042003) and their corresponding controls were obtained from RiboBio. The oligonucleotides were transfected into U251 and U251R cells using Lipofectamine 3000 reagent (Invitrogen) following the manufacturer’s protocols.
2.3. Cell counting kit‐8 assay
Cell Counting Kit‐8 (Beyotime) was used to evaluate the IC50 value of TMZ. In brief, glioma cells (2 × 103) were incubated in media containing different concentrations of TMZ (0, 0.1, 1, 5, 10, 20, 100, 200, and 400 μM) for 48 hours. Ten microliters of CCK‐8 solution (Beyotime) was added to each well and incubated for 2 hours. The absorbance at 450 nm was measured with a microplate reader. GraphPad Prism version 8.0 (GraphPad Software) was used to calculate the IC50 of TMZ. For exosome treatment analysis, 2 × 103 cells were seeded into 96‐well plates, and exosomes were loaded at a concentration of 10 μg/mL. After incubating at 37℃ for 48 hours, the medium was removed, and fresh medium containing different concentrations of TMZ was added to each well and cells were further cultured for 48 hours. At the end of the treatment, the absorbance at 450 nm was determined, and IC50 was calculated.
For cell proliferation determination, U251 and U251R cells (2 × 103) were seeded into 96‐well plates, and cell growth was determined by CCK‐8 assay every 24 hours for three consecutive days.
2.4. 5‐Ethynyl‐2′‐deoxyuridine incorporation assay
After exosome loading (or not), cells were cultured with 30 μM TMZ for 48 hours. Cell proliferation was also assessed by the 5‐ethynyl‐2′‐deoxyuridine (EdU) assay using a Cell‐Light EdU DNA Cell Proliferation Kit (RiboBio) according to the instructions.
2.5. Flow cytometry
The apoptosis rate after TMZ treatment was determined by flow cytometry. After 48 hours of incubation with 30 μM TMZ, the cells were collected and labeled with annexin V‐FITC and propidium iodide (BD Biosciences). Apoptotic cells were quantified by a fluorescence‐activated cell sorting flow cytometer (BD Biosciences).
2.6. Quantitative real‐time PCR
Total RNA of exosomes from serum was isolated using the exoRNeasy serum/plasma midi kit (Qiagen), and total RNA from cells and exosomes derived from cells was isolated using TRIzol reagent (Invitrogen). The complementary DNA was synthesized by the High‐Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The SYBR Green PCR Master Mix (Thermo Fisher Scientific) was mixed with primers for the quantitative real‐time PCR (qRT‐PCR) assay. The qRT‐PCR was undertaken using an ABI 7500 real‐time PCR system (Applied Biosystems). Gene expression was calculated using the 2−ΔΔCt method; GAPDH was used as the internal control. The primers are displayed in Table 1.
TABLE 1.
Primer sequences used this study
| Gene | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| Heparanase | 5′‐CCC TTG CTA TCC GAC ACC TT‐3′ | 5′‐CAC CAC TTC TAT TCC CTT TCG‐3′ |
| hsa_circ_0041638 | 5′‐CCG TGT GTA AGG CGG GAG‐3′ | 5′‐CAT AGA TAC CCG TGG AAG CCC‐3′ |
| hsa_circ_0005963 | 5′‐GGC ATG TGT GGT CGT TTT TCA‐3′ | 5′‐AGG TCA CAA CAA TGG ATG CC‐3′ |
| hsa_circ_0037096 | 5′‐GTC CGC AAC CCG GAG AAG‐3′ | 5′‐CCA CCA CGT GCT CCA AGT AG‐3′ |
| hsa_circ_0002782 | 5′‐ACC TGC CAT CAA TGG TGT GA‐3′ | 5′‐GCT GTA AGA CTG CTG GAC AGA‐3′ |
| hsa_circ_0042003 | 5′‐CCA AGA TGT GCT GGA TGC AA‐3′ | 5′‐CTA CTG CTC CAC GAC AGG TG‐3′ |
| GAPDH | 5′‐GTC AAC GGA TTT GGT CTG TAT T‐3′ | 5′‐AGT CTT CTG GGT GGC AGT GAT‐3′ |
2.7. Western blot assay
Total proteins were exacted from cells and exosomes using lysis buffer (Beyotime). The protein concentration was analyzed using a BCA Protein Assay Kit (Beyotime). Proteins were separated using SDS‐PAGE and then transferred onto PVDF membranes (Roche Applied Science). The membranes were blocked with 2% BSA (Sigma‐Aldrich), followed by incubation with the indicated primary Abs overnight at 4℃. Membranes were then incubated with HRP‐conjugated goat anti‐mouse or goat anti‐rabbit IgG for 2 hours at room temperature, and the immunoblots were visualized using an enhanced chemiluminescence kit (GE Healthcare). The following primary Abs were used: anti‐heparanase (sc‐25825; Santa Cruz Biotechnology), anti‐CD63 (ab134045, 1:1000; Abcam), anti‐CD9 (ab92726, 1:1000; Abcam), anti‐CD81 (ab219209, 1:1000; Abcam), anti‐Flotillin‐1 (ab133497, 1:10 000; Abcam), and anti‐β‐actin (Abcam).
2.8. Isolation and characterization of exosomes
As previously described,20 exosomes were isolated from vesicle‐depleted medium and serum samples using the Total Exosome Isolation Kit (Thermo Fisher Scientific). The exosomal protein concentration was detected by BCA colorimetric method. The morphology of the exosomes was observed using transmission electron microscopy (JEM1011 Microscope; Jeol),21 and the size of exosomes was determined by the Nanoparticle Tracking Analysis (NTA) using NanoSight NS300 (NanoSight Technology).22 The protein markers of exosomes were identified using western blot assay, and anti‐flotillin‐1, anti‐CD63, anti‐CD9, and anti‐CD81 Abs were used.
2.9. Circular RNA microarray
Exosomal RNA from cell culture media was isolated using the Qiagen exoEasy Maxi Kit (Qiagen). Total RNA was digested with RNase R. The quality of RNA was measured by NanoDrop ND‐1000 (NanoDrop Technologies). The Arraystar Flash RNA Labeling Kit (Arraystar) was used to amplify enriched circRNAs and transcribed them into fluorescent cRNA. After purification by the RNeasy Mini Kit (Qiagen), the labeled cRNAs were hybridized to Arraystar circRNA Microarray version 2.0. The G2565CA microarray scanner system (Agilent Technologies) was used for microarray scanning. Arraystar software was used to normalize the microarray data and the differentially expressed circRNAs (P < .05 and fold change greater than 2) were identified. Hierarchical clustering was carried out to visualize the circRNAs.
2.10. PKH26 staining
Purified exosomes were labeled using a PKH26 red fluorescent labeling kit (Sigma‐Aldrich). Exosomes (300 μL) were resuspended in 100 μL Diluent C, and another 300 μL Diluent C was mixed with 1.4 μL PKH26 to prepare the staining solution. The exosome suspension was mixed with the staining solution and incubated for 5 minutes. The labeled exosomes were added into U251 or U251R cells, and then incubated at 37℃ for 6 hours. The cells were fixed with 4% paraformaldehyde, and then stained with DAPI. An FV1000 confocal microscope (Olympus) equipped with the UPlanSApo 40×/0.75 objective lens with red fluorescence intensity was used to determine the uptake of the labeled exosomes by recipient cells.
2.11. Xenograft model
Female BALB/c nude mice (6 weeks old) were obtained from Beijing HFK Bioscience and housed in a special pathogen‐free animal facility. Animal studies were approved by the Ethics Committee of The Second Affiliated Hospital of Zhengzhou University. In order to evaluate the role of exosomes in vivo, exosomes derived from U251R cells were collected. Nude mice were randomly selected and divided into two groups, the PBS group and U251R exo group, with five animals in each. U251 cells (2 × 106) were injected subcutaneously in the right flank. When the tumor volume exceeded 500 mm3 (15 days after inoculation), the PBS group was injected with 30 μL PBS daily through the tail vein, while U251R exo group was given 15 μg U251R cell‐derived exosomes (exosomes were resuspended in 30 μL PBS). At the same time, TMZ (40 mg/kg) was given daily by intraperitoneal injection. The tumor size was quantified every 4 days according to the formula: tumor volume (mm3) = (shortest diameter)2 × (longest diameter) × 0.52. After 28 days of treatment, blood and xenograft tumors of nude mice were collected for subsequent determination. Immunohistochemistry (IHC) analysis was carried on mice xenogeneic tumor tissues as described previously.23
In order to evaluate whether exosomes work by exosomal hsa_circ_0042003 or heparanase in vivo, exosomes derived from U251 cells transfected with si‐control (si‐control exo), si‐heparanase (si‐heparanase exo), or si‐hsa_circ_0042003 (si‐hsa_circ_0042003 exo) were collected. First, U251R cells (2 × 106) were subcutaneously inoculated into the right flank of 6‐week‐old nude mice. When the smallest xenograft tumor volume was approximately 500 mm3, the nude mice were randomly divided into four groups, PBS, si‐control exo, si‐heparanase exo, and si‐hsa_circ_0042003, with five animals in each group. Then nude mice were cotreated with intraperitoneal injection of TMZ (40 mg/kg) and tail injection of 15 μg exosomes (si‐control exo, si‐heparanase exo, or si‐hsa_circ_0042003 exo) or PBS daily. The tumor volume was measured and calculated every 4 days. The blood and xenograft tumor of nude mice were collected after 28 days of continuous treatment.
2.12. Statistical analysis
Statistical analyses were undertaken using GraphPad Prism version 8.0 (GraphPad Software). Data are shown as mean ± SD of at least three independent experiments. Student’s t test or ANOVA was used for comparison between groups. P < .05 was considered to be statistically significant.
3. RESULTS
3.1. Expression of heparanase in glioma cells correlates with their resistance to TMZ
The IC50 values of TMZ in glioma cells are shown in Figure 1A. Among them, U251 had the highest sensitivity to TMZ. Heparanase mRNA expression was positively correlated with the IC50 of glioma cells to TMZ (Figure 1B). The IC50 of U251R cells (160.4 μM) was sharply higher than that of the U251 parental cells (16.38 μM), suggesting a successful establishment of TMZ‐resistant glioma cells (Figure 1C). Temozolide resistance affected the expression of heparanase, including promoting the expression of heparanase mRNA (Figure 1D) and protein (Figure 1E). Compared with U251 cells, U251R cells showed increased cell viability (Figure 1F). Together, the changes in cell activity induced by TMZ resistance might be related to the upregulated expression of heparanase.
FIGURE 1.
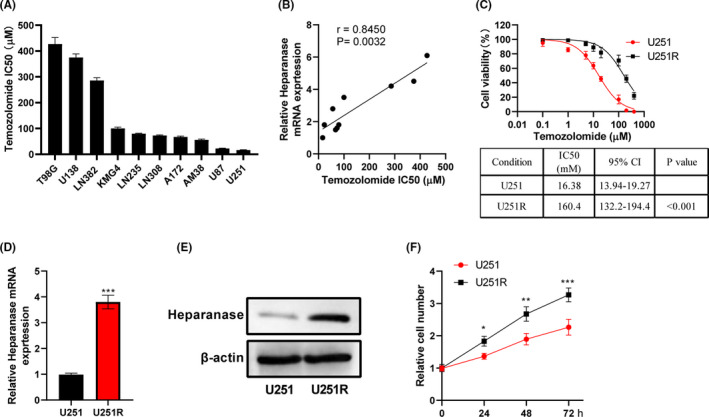
Expression of heparanase in glioma cells correlates with their resistance to temozolomide (TMZ). A, CCK‐8 assay was used to analyze the viability of glioma cells treated with different concentrations of TMZ (0, 0.1, 1, 5, 10, 20, 100, 200, 400 μM) for 48 hours, and the IC50 of TMZ was calculated. B, The relationship between heparanase mRNA and the IC50 of TMZ in glioma cells exposed to TMZ was analyzed. C, Viability of U251 and U251R cells exposed to TMZ was analyzed using CCK‐8 assay. CI, confidence interval. D, The expression of heparanase mRNA in U251R cells was markedly higher than that of U251 cells. E, Western blot assay was used to analyze the expression of heparanase in U251 and U251R cells. F, Proliferation of U251 and U251R cells was analyzed using CCK‐8 assay. *P < .05; **P < .01; ***P < .001
3.2. Heparanase confers resistance to TMZ in glioma cells
As shown in Figure 2A, overexpression of heparanase increased the IC50 value of TMZ in U87 cells, suggesting upregulation of heparanase could increase the resistance of TMZ‐sensitive glioma cells to TMZ. Additionally, knockdown of heparanase decreased the IC50 value of TMZ in U251R cells, suggesting downregulation of heparanase could increase the sensitivity of TMZ‐resistant glioma cells to TMZ (Figure 2B). Moreover, EdU assay showed that knockdown of heparanase inhibited cell proliferation of U251R cells treated with TMZ (Figure 2C). Inhibition of apoptosis is one of the manifestations related to TMZ resistance. It was found that knockdown of heparanase induced apoptosis of U251R cells exposed to TMZ (Figure 2D). These data revealed that heparanase enhanced glioma cells’ resistance to TMZ.
FIGURE 2.

Heparanase confers resistance to temozolomide (TMZ) in glioma cells. A, The viability of U87 cells transfected with heparanase pcDNA was analyzed using CCK‐8 assay when exposed to different concentration of TMZ. B, The viability of U251R cells transfected with heparanase siRNA was analyzed using CCK‐8 assay when exposed to different concentration of TMZ. C, Proliferation of U251R cells transfected with heparanase siRNA was detected using EdU assay when exposed to 30 μM TMZ. Scale bar = 50 μM. D, Apoptosis of U251R cells transfected with heparanase siRNA was detected using flow cytometry when exposed to 30 μM TMZ. ***P < .001. CI, confidence interval
3.3. Heparanase promotes exosome secretion
As shown in Figure 3A, U251R cells secreted more exosomes than U251 cells. To investigate the role of heparanase on the secretion of exosomes, si‐heparanase was transfected into U251R cells. As expected, knockdown of heparanase reduced the secretion of exosomes from U251R cells (Figure 3A). As shown in Figure 3B, flotillin‐1, CD63, CD9, and CD81 were more enriched in exosomes derived from U251R cells than U251 cells, and knockdown of heparanase reduced flotillin‐1, CD63, CD9, and CD81 levels in U251R‐derived exosomes. Transmission electron microscopy revealed that the vesicles isolated from U251 and U251R cells contained a round or oval membrane (Figure 3C). The NTA analysis showed that most of the vesicles had a diameter in the range of 50–150 nm (data no shown). Therefore, heparanase might promote the secretion of exosomes derived from glioma cells.
FIGURE 3.
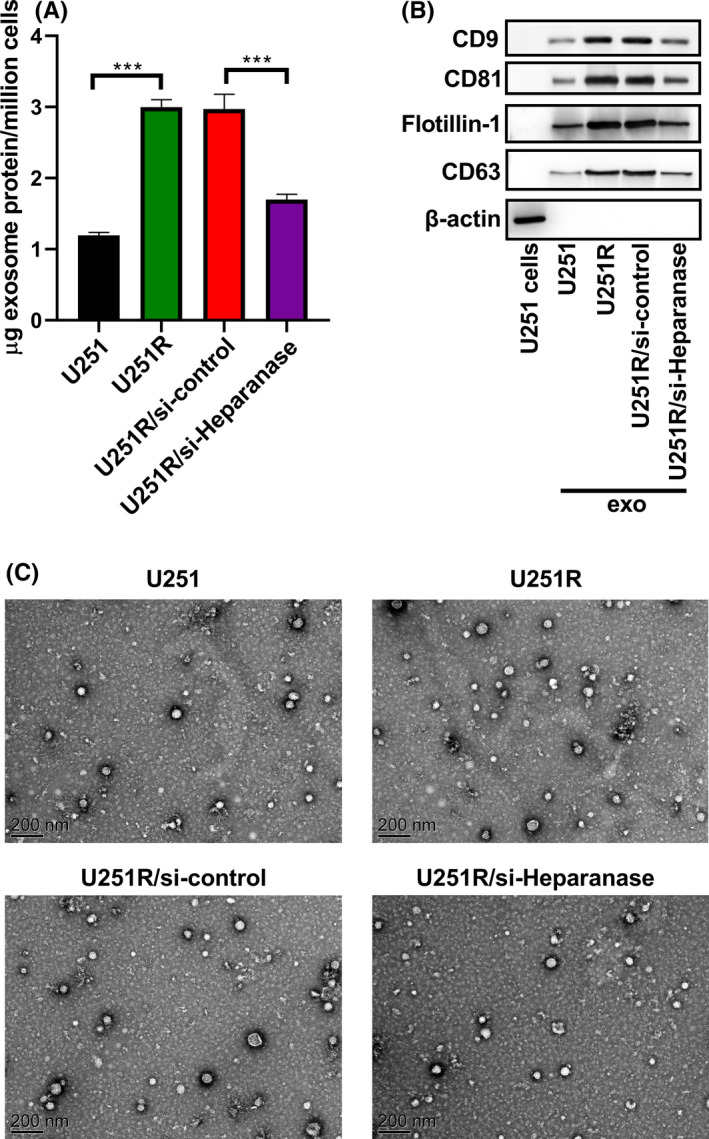
Heparanase promotes exosome secretion. A, Content of exosome protein isolated from U251 and U251R cells transfected with or without heparanase siRNA was detected by the BCA colorimetric method. B, Expression of exosome (exo) marker proteins flotillin‐1, CD63, CD9, and CD81 was analyzed using western blot assay. C, Morphology of exosomes was observed using transmission electron microscopy. ***P < .001
3.4. U251R cell‐derived exosomes enhance TMZ resistance in parental U251 cells
Exosomes have been identified to be related to chemoresistance of cancer cells.24 Therefore, we investigated whether exosomes function in TMZ resistance in glioma cells. U251 cells were cocultured with exosomes derived from U251R and U251 cells, respectively, before TMZ treatment. As shown in Figure 4A, immunofluorescence confirmed that PKH26‐labeled exosomes derived from U251 and U251R cells could be delivered to U251 cells. Additionally, coculture with exosomes derived from U251R cells could enhance the resistance of U251 cells to TMZ, which was manifested by a significantly increased IC50 value (Figure 4B). Knockdown of heparanase attenuated the enhancement of U251R cell‐derived exosomes on the chemoresistance of U251 cells (Figure 4B). Temozolide‐resistant cell‐derived exosomes led to a marked enhancement in the proliferation, and clearly weakened apoptosis, of U251 cells (Figure 4C,D). Simultaneously, heparanase knockdown in exosomes derived from U251R cells could rescue the effects of TMZ‐resistant cell‐derived exosomes on the proliferation and apoptosis of U251 cells. These results confirmed that exosomes derived from TMZ‐resistant cells could reduce the sensitivity of U251 cells to TMZ, and heparanase was involved in this process.
FIGURE 4.
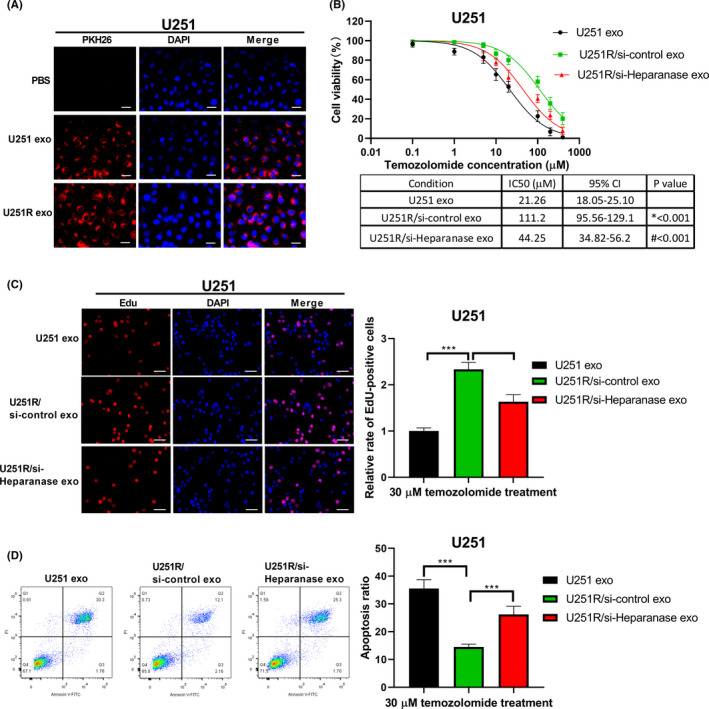
U251R cell‐derived exosomes enhance resistance to temozolomide (TMZ) in parental U251 cells. A, PKH26‐labeled exosomes (exo) derived from U251 and U251R cells could fuse into U251 cells. Scale bar = 20 μM. B, U251 cells were cocultured with exosomes derived from U251R cells transfected with or without heparanase siRNA, and then exposed to different concentrations of TMZ. Viability of U251 cells was detected using CCK‐8 assay. CI, confidence interval. C, Proliferation of U251 cells was analyzed using EdU assay. Scale bar = 50 μM. D, Apoptosis of U251 cells was analyzed using flow cytometry. ***P < .001
3.5. Hsa_circ_0042003 is upregulated in U251R cell‐derived exosomes and serum exosomes of TMZ‐resistant glioma patients
Circular RNAs can be carried by exosomes and transferred to adjacent or distant cells for intercellular communication.22 Therefore, we undertook a microarray using exosomes derived from U251 and U251R cells. Compared with exosomes derived from U251 cells, 104 circRNAs changed significantly in the exosomes derived from U251R cells (P < .05 and fold change greater than 2), of which 55 were upregulated and 49 were downregulated (Figure 5A). The top five most increased circRNAs (hsa_circ_0041638, hsa_circ_0005963, hsa_circ_0037096, hsa_circ_0002782, and hsa_circ_0042003; Table 2) were selected for verification in exosomes derived from U251 and U251R cells (Figure 5B), or exosomes derived from serum of patients with and without TMZ‐resistant glioma (Figure 5C) using qRT‐PCR. There were no significant differences in the baseline clinical characteristics between TMZ‐resistant and TMZ‐sensitive patients (Table 3). The results of qRT‐PCR revealed that, in equal exosomal protein amounts, hsa_circ_0042003 was significantly increased in U251R cell‐derived exosomes compared with exosomes derived from U251 cells (Figure 5B). Simultaneously, hsa_circ_0042003 in serum exosomes derived from TMZ‐nonresponders was markedly higher than those derived from TMZ‐responders (Figure 5C).
FIGURE 5.
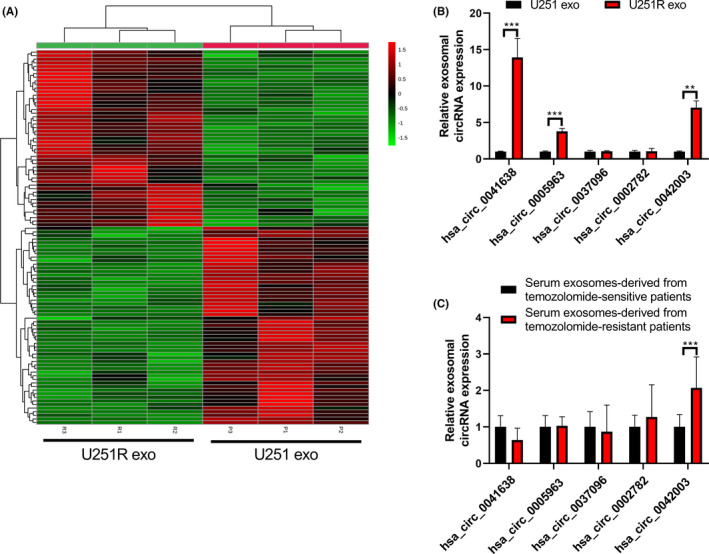
Hsa_circ_0042003 is upregulated in exosomes derived from U251R cells and the serum exosomes of temozolomide (TMZ)‐resistant glioma patients. A, Hierarchical clustering was used to identify differentially expressed exosomal circRNAs derived from U251 and U251R cells. B, Expression of hsa_circ_0041638, hsa_circ_0005963, hsa_circ_0037096, hsa_circ_0002782, and hsa_circ_0042003 in exosomes (exo) derived from U251 and U251R cells was analyzed using quantitative real‐time PCR (qRT‐PCR). C, Expression of hsa_circ_0041638, hsa_circ_0005963, hsa_circ_0037096, hsa_circ_0002782, and hsa_circ_0042003 in exosomes derived from serum of patients with and without TMZ‐resistant glioma was analyzed using qRT‐PCR. **P < .01; ***P < .001
TABLE 2.
Top five upregulated circular RNAs (circRNAs) in U251R cells
| CircRNA | Gene | Fold change | Log2(FC) | P value | Chr | Type |
|---|---|---|---|---|---|---|
| hsa_circ_0041638 | ENO3 | 23.965 | 4.5829 | 0.0019356 | 17 | Exons |
| hsa_circ_0005963 | TMEM128 | 16.024 | 4.0022 | 0.0026448 | 4 | Exons |
| hsa_circ_0037096 | PCSK6 | 13.915 | 3.7986 | 0.0041371 | 15 | Exons |
| hsa_circ_0002782 | SLC39A8 | 12.789 | 3.6768 | 0.0224070 | 4 | Exons |
| hsa_circ_0042003 | CTC1 | 10.488 | 3.3907 | 0.0172010 | 17 | Exons |
Abbreviations: Chr, chromosome number; FC, fold change.
TABLE 3.
Baseline clinical characteristics of 40 glioma patients
| Characteristic | All patients | TMZ nonresponders | TMZ responders | P value |
|---|---|---|---|---|
| Gender | ||||
| Male | 25 | 12 | 13 | >0.05 |
| Female | 15 | 8 | 7 | |
| Age (y) | ||||
| ≥50 | 25 | 11 | 14 | >0.05 |
| <50 | 15 | 9 | 6 | |
| KPS score | ||||
| ≥80 | 7 | 3 | 4 | >0.05 |
| <80 | 33 | 17 | 16 | |
| Tumor size (cm) | ||||
| ≥4 | 22 | 12 | 10 | >0.05 |
| <4 | 18 | 8 | 10 | |
Abbreviation: KPS, Karnofsky performance status.
3.6. Heparanase regulates TMZ resistance in glioma cells by enhancing hsa_circ_0042003 expression
As shown in Figure 6A, si‐heparanase transfection could markedly reduce the expression of hsa_circ_0042003 in exosomes derived from U251R cells. Additionally, heparanase pcDNA transfection promoted hsa_circ_0042003 expression in exosomes derived from U251 cells (Figure 6B). Moreover, si‐heparanase transfection decreased hsa_circ_0042003 expression in U251R cells (Figure 6C), whereas pcDNA‐heparanase transfection increased hsa_circ_0042003 expression in U251 cells (Figure 6D). These data revealed that heparanase positively regulated hsa_circ_0042003 expression and participated in the mediation of hsa_circ_0042003 from cells to exosomes. Compared with coculture with U251 cell‐derived exosomes, coculture with exosomes derived from U251R cells significantly increased the level of hsa_circ_0042003 in U251 cells (Figure 6E), suggesting that hsa_circ_0042003 could be received by recipient cells through exosomes.
FIGURE 6.
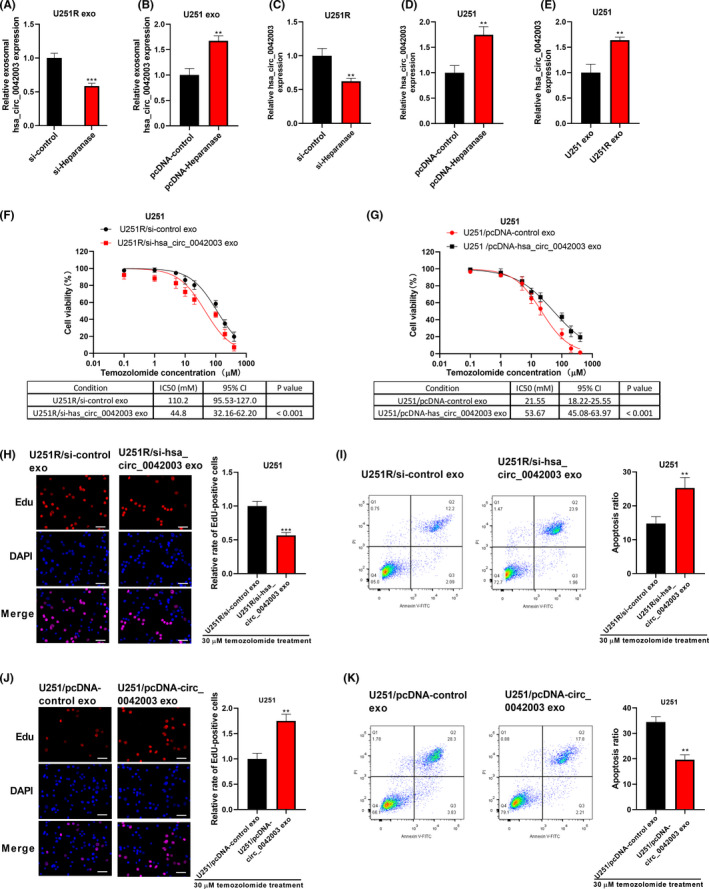
Heparanase regulates temozolomide (TMZ) resistance in glioma cells by enhancing hsa_circ_0042003 expression. Exosomes were isolated from U251R cells transfected with si‐heparanase, or U251 cells transfected with pcDNA‐heparanase. A, B, Expression of exosomal hsa_circ_0042003 was analyzed using quantitative real‐time PCR (qRT‐PCR). C, After U251R cell transfection with si‐heparanase, hsa_circ_0042003 expression in U251R cells was detected using qRT‐PCR. D, After U251 cell transfection with pcDNA‐heparanase, hsa_circ_0042003 expression in U251 cells was detected using qRT‐PCR. E, U251 cells were cocultured with exosomes (exo) derived from U251 or U251R cells, and then hsa_circ_0042003 expression in U251 cells was detected using qRT‐PCR. U251 cells were cocultured with exosomes derived from U251R cells transfected with si‐hsa_circ_0042003, or U251 cells transfected with pcDNA‐hsa_circ_0042003. F, G, CCK‐8 assay was used to evaluate the viability of U251 cells. H, J, Proliferation of U251 cells was evaluated using EdU assay. Scale bar = 50 μM. I, K, Apoptosis of U251 cells was analyzed using flow cytometry. **P < .01; ***P < .001
To assess whether hsa_circ_0042003 was related to TMZ resistance, U251 cells were cocultured with exosomes derived from U251R cells transfected with si‐hsa_circ_0042003 or exosomes derived from U251 cells transfected with pcDNA‐hsa_circ_0042003. In a series of functional experiments, U251R cell‐derived exosomal si‐hsa_circ_0042003 could abolish the TMZ resistance of U251 cells induced by control U251R‐derived exosomes (Figure 6F), resulting in decreased proliferation (Figure 6H) and increased apoptosis (Figure 6I). In contrast, U251 cell‐derived exosomal pcDNA‐hsa_circ_0042003 increased the resistance of U251 cells to TMZ (Figure 6G, J, K).
3.7. Exosomes derived from chemoresistant cells decrease the drug susceptibility of chemosensitive cells in vivo
Although it is still unclear how drug‐sensitive cells develop chemoresistance, some studies have suggested that this might involve the intercellular communication function of exosomes.25 Considering that sensitive cells could be affected by exosomes derived from drug‐resistant cells, we divided nude mice into a PBS group and a U251R exo group. A xenograft model was established according to the process shown in Figure 7A. The exosomes derived from U251R cells were collected and characterized (Figure 7B,C). U251 cells (2 × 106) were injected subcutaneously in the right flank. When the tumor volume exceeded 500 mm3, the PBS group was injected with 30 μL PBS daily through the tail vein, while the U251R exo group was given 15 μg U251R‐derived exosomes. At the same time, TMZ (40 mg/kg) was given daily by intraperitoneal injection. The tumor size was quantified every 4 days. After 28 days of treatment, blood and xenograft tumors of nude mice were collected. The volume of xenograft tumors in the U251R exo group was significantly higher than that in the PBS group (Figure 7D). Subsequently, we evaluated whether U251R‐derived exosomes affected TMZ resistance of U251 cells through exosomal hsa_circ_0042003. As shown in Figure 7E,G, the U251R exo group had significantly higher hsa_circ_0042003 levels in serum exosomes and tumors than the PBS group. Similar to the change in tumor volume, xenograft tumor weight also revealed that, compared with U251 cells in the PBS group, the proliferation ability of U251 cells in the U251R exo group was significantly less inhibited by TMZ treatment (Figure 7F). In addition, IHC analysis showed that xenograft tumor in the U251R exo group had higher proliferating cell nuclear antigen (PCNA) expression and lower caspase‐3 expression than the PBS group (Figure 7H). These data revealed that U251R cell‐derived exosomes could enhance the resistance of U251 cells to TMZ, which might involve the process of exosomes delivering hsa_circ_0042003 to sensitive cells.
FIGURE 7.
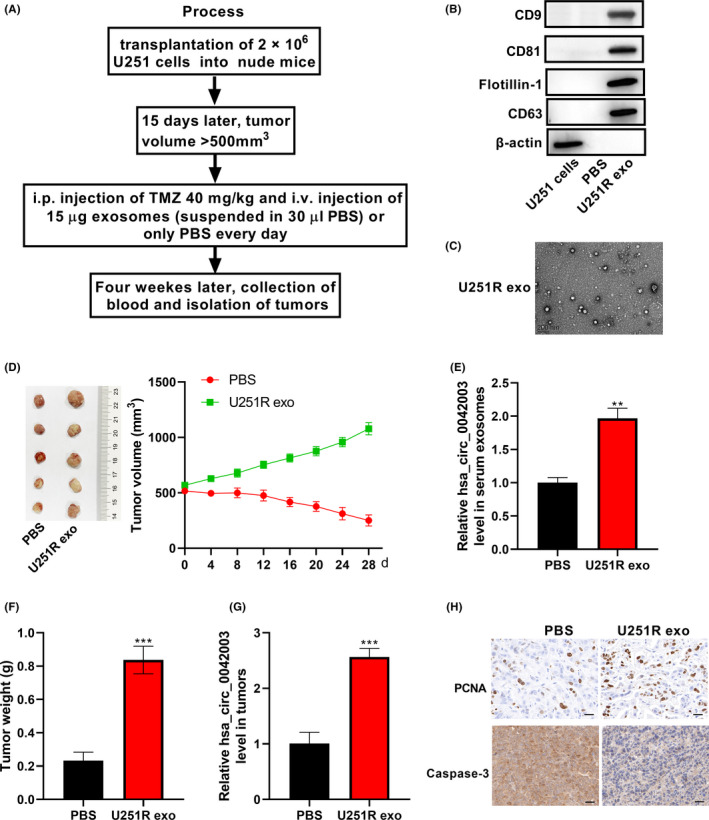
Exosomes derived from chemoresistant cells decrease the drug susceptibility of chemosensitive cells in vivo. A, Experimental design diagram of the xenograft model. Exosomes derived from U251R cells were collected. B, Expression of exosome (exo) marker proteins flotillin‐1, CD63, CD9, and CD81 was analyzed using western blot assay. C, Morphology of exosomes was observed using transmission electron microscopy. D, Volume of xenograft tumors changed during TMZ and exosome treatment. After 28 days of treatment, mouse serum was collected and serum exosomes were extracted. E, Quantitative real‐time PCR (qRT‐PCR) was used to assess the level of hsa_circ_0042003 in serum exosomes. F, Tumors were resected from mice at the same time on day 28 of treatment, and the tumor weight is shown. G, Level of hsa_circ_0042003 in xenograft tumors was analyzed using qRT‐PCR. H, Immunohistochemical analysis of the expression of proliferating cell nuclear antigen (PCNA) and caspase‐3 in xenograft tumors. Scale bar = 20 μM. **P < .01; ***P < .001
3.8. Systemically injected si‐heparanase or si‐hsa_circ_0042003 exosomes could sensitize the response to TMZ in vivo
Subsequently, the effects of exosomal heparanase and exosomal hsa_circ_0042003 on the growth of TMZ‐resistant glioma cells were investigated in vivo. In view of the strong proliferation ability of U251R cells, subcutaneous injection of U251R cells was chosen to construct a nude mouse model. When the tumors had grown to a suitable size, 15 μg of different exosomes (resuspended in 30 μL PBS) or PBS was injected through the tail vein combined with intraperitoneal injection of TMZ (40 mg/kg). Over time, the tumors in the PBS group and si‐control exo group progressed, while the tumors in the si‐heparanase exo and si‐hsa_circ_0042003 exo groups were stable (Figure 8A). Exosomes were isolated from nude mouse serum and hsa_circ_0042003 levels were determined using qRT‐PCR. The si‐heparanase exo and si‐hsa_circ_0042003 exo groups had significantly reduced hsa_circ_0042003 levels (Figure 8B). Weights of the xenograft tumors revealed that si‐heparanase exo and si‐hsa_circ_0042003 exo resulted in significantly smaller tumors (Figure 8C). In addition, the expression of hsa_circ_0042003 in xenograft tumors of the si‐heparanase exo group and the si‐hsa_circ_0042003 exo group was significantly reduced (Figure 8D). The IHC analysis showed that si‐heparanase exo and si‐hsa_circ_0042003 exo groups had significantly decreased PCNA expression and increased caspase‐3 expression (Figure 8E). Our findings suggested that heparanase accelerated the transmission of exosomal hsa_circ_0042003 from TMZ‐resistant glioma cells to drug‐sensitive cells, thereby promoting the chemoresistance of glioma to TMZ (Figure 9). Inhibition of heparanase‐mediated transmission of hsa_circ_0042003‐rich exosomes might be a possible mechanism to enhance the sensitivity of drug‐resistant cells to TMZ in vivo.
FIGURE 8.
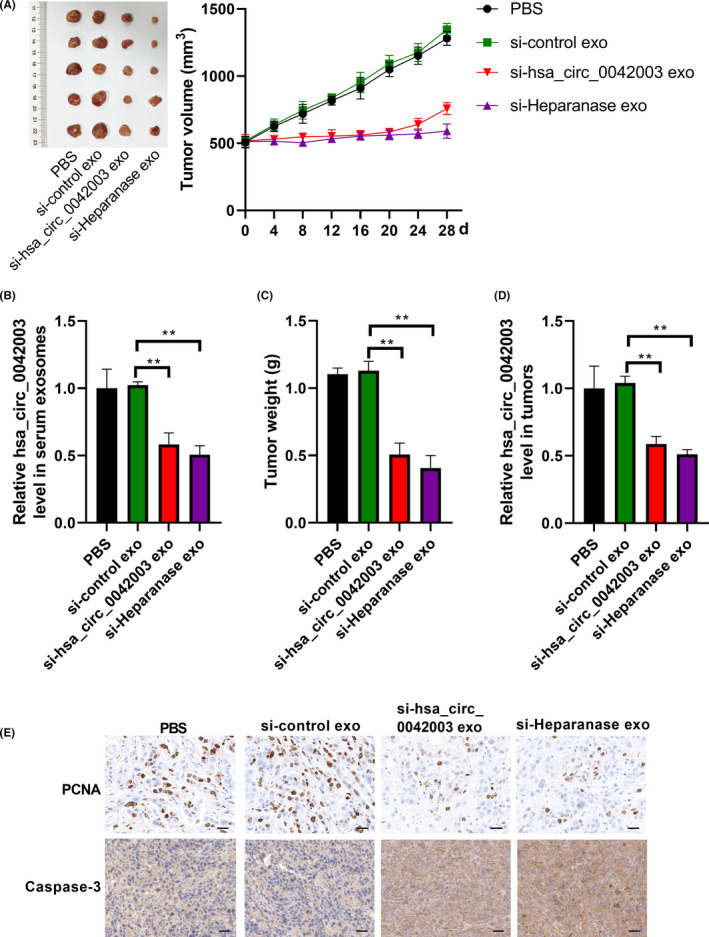
Systemically injected si‐heparanase or si‐hsa_circ_0042003 exosomes could sensitize the response to temozolomide (TMZ) in vivo. When the volume of tumors developed by inoculated U251R cells exceeded 500 mm3, exosomes derived from U251 cells transfected with si‐control (si‐control exo), si‐heparanase (si‐heparanase exo), or si‐hsa_circ_0042003 (si‐hsa_circ_0042003 exo) combined with TMZ were used to treat transplanted tumors in a nude mouse model. A, Tumor volume was recorded every 4 days during treatment. After 28 days of treatment, serum from nude mice was collected and serum exosomes were separated. B, Level of hsa_circ_0042003 in serum exosomes was detected using quantitative real‐time PCR (qRT‐PCR). C, Tumors were resected from mice at the same time on day 28 of treatment, and the actual tumors and tumor weight are shown. D, Levels of hsa_circ_0042003 in xenograft tumors were analyzed using qRT‐PCR. E, Immunohistochemical analysis of the expression of proliferating cell nuclear antigen (PCNA) and caspase‐3 in xenograft tumors. Scale bar = 20 μM. **P < .01; ***P < .001
FIGURE 9.
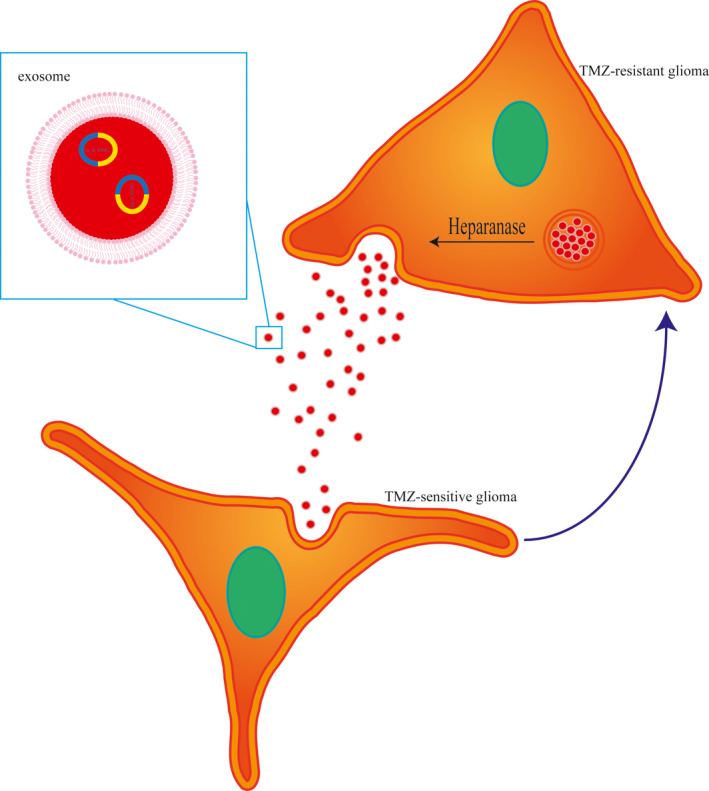
Schematic illustration of the mechanism underlying the role of heparanase in temozolomide (TMZ) resistance in glioma
4. DISCUSSION
Chemoresistance is one of the main problems in the management of glioma, which seriously affects the prognosis of patients. It is necessary to study the related mechanisms of TMZ resistance and determine new targets for the prevention and therapy of TMZ resistance. Exosomes secreted by tumor cells have become a new hotspot in the research of chemoresistance,16, 26 and heparanase has also been confirmed to be involved in mediating the secretion and function of exosomes.18 In this study, we found that heparanase was positively correlated with the resistance of glioma cells to TMZ, which might be achieved by mediating the intercellular communication function of exosomes. Additionally, we identified an exosomal circRNA related to TMZ resistance, hsa_circ_0042003, which might be involved in the role of heparanase in TMZ resistance. The nude mouse xenograft model also confirmed that exosomal hsa_circ_0042003‐mediated communication between cells might be the mechanism that causes drug‐resistant cells to induce TMZ resistance in sensitive cells. Moreover, the combination of tail‐injected exosomal si‐hsa_circ_0042003 or si‐heparanase and intraperitoneal TMZ applied to nude mice abolished chemoresistance in vivo. It can be concluded from this study that heparanase could facilitate tumor resistance by promoting the delivery of exosomal hsa_circ_0042003.
Resistance is currently the biggest obstacle to glioma therapy. It has been confirmed that targeting heparanase can inhibit the progression and chemoresistance of a variety of tumors.9, 12, 27 In this study, we also confirmed that heparanase is positively correlated with TMZ resistance in glioma. Simultaneously, inhibition of heparanase restrained a TMZ‐resistant phenotype and enhanced TMZ‐induced cell death in TMZ‐resistant glioma cells. Additionally, in the nude mouse model established by inoculation with TMZ‐resistant cells, the depletion of heparanase in exosomes showed enhanced TMZ sensitivity compared with empty exosomes. These results revealed that targeting heparanase could benefit the treatment of glioma patients receiving TMZ.
Initially, drug‐resistant and sensitive tumor cells coexist, but most of the cells eventually become drug‐resistant.28 How does this happen? As a key mediator of communication between tumor cells or other cells in the microenvironment, exosomes secreted by tumor cells have become a new model for chemoresistance research.25 Exosomes derived from chemoresistant cancer cells modify the cell phenotype to confer drug resistance on sensitive cells.29, 30 For instance, exosomes isolated from cisplatin‐resistant breast cancer cells can enhance the resistance of breast cancer cells to cisplatin.31 In the current study, we also confirmed that TMZ‐resistant glioma cell‐derived exosomes could confer TMZ resistance to the parental sensitive glioma cells. It has been reported that the activity of heparanase is necessary to enhance the secretion of exosomes.18 Consistently, the knockdown of heparanase in the current study reduced the secretion of exosomes in TMZ‐resistant glioma cells in vitro. In addition, inhibition of heparanase enhanced the sensitivity of resistant cells to TMZ, which might be achieved by weakening the intercellular communication function of exosomes. It was also observed in the nude mouse xenograft model that knockdown of heparanase in exosomes reduced TMZ resistance in resistant cells in vivo. Therefore, inhibition of heparanase could regulate the influence of drug‐resistant cells on sensitive cells by reducing the transfer and communication of exosomes between cells.
The proteins and genetic material carried by exosomes play an important role in the signal transduction between cells.32, 33 For example, exosomes transfer the migration inhibitory factor protein to the Kupffer immune cells located in the liver, thereby promoting the metastasis of primary cancer cells.34 It has recently been confirmed that circRNAs play a key role in cancer progression,35 especially in chemoresistance.36 Interestingly, exosomal circRNAs have also been found to be involved in the signal exchange between tumor cells, the local microenvironment, and remote target organs.37 Here, we used a microarray analysis and identified the high expression of hsa_circ_0042003 in TMZ‐resistant glioma cell‐derived exosomes. The high expression of hsa_circ_0042003 was confirmed in exosomes derived from serum of glioma patients with TMZ resistance. Our results showed that downregulation of hsa_circ_0042003 abolished the TMZ resistance of sensitive cells induced by TMZ‐resistant cell‐derived exosomes in vitro, while the acquisition of hsa_circ_0042003 increased the TMZ resistance of sensitive cells. In addition, in vivo experiments also proved that knockdown of exosomal hsa_circ_0042003 partially eliminated the TMZ resistance of drug‐resistant cells. These findings suggested that blocking the internalization of exosomal hsa_circ_0042003 might be an effective strategy to reduce the chemoresistance of glioma cells.
Heparanase can enhance the secretion of exosomes interacting with tumors and host cells, and promote their development toward aggressive tumor phenotypes,38 which involves the influence of heparanase on the composition of exosomes, including syndecan‐1, vascular endothelial growth factor, and hepatocyte growth factor.18 However, no studies have reported whether heparanase can regulate the cargo of circRNAs in exosomes. Interestingly, we found that the level of hsa_circ_0042003 was positively regulated by heparanase, which suggested that heparanase‐mediated TMZ resistance might involve hsa_circ_0042003. Previous studies have confirmed that circRNA carried by exosomes derived from drug‐resistant cells is absorbed by drug‐sensitive cells to promote drug resistance.26, 39 Therefore, drug‐resistant cell‐derived exosomes could increase the resistance of sensitive cells to TMZ in vivo, which could involve in the process of exosomes delivering hsa_circ_0042003 to sensitive cells. In addition, exosomal heparanase knockdown enhanced the sensitivity of drug‐resistant cells to TMZ in vivo, which decreased hsa_circ_0042003 expression in serum exosomes and tumors. Collectively, targeting heparanase to regulate exosome‐mediated hsa_circ_0042003 delivery could become a potential method to prevent and treat TMZ resistance. In addition, circRNAs have been widely reported to function by acting as microRNA sponges. Increasing evidence has revealed that circRNAs regulate chemoresistance by sponging microRNAs.40, 41 Given these findings, we intend to further explore whether heparanase‐hsa_circ_0042003 functions through microRNAs.
In conclusion, the present study showed that heparanase contributed to the resistance of glioma to TMZ, which involved its promotion of the secretion of TMZ‐resistant cell‐derived exosomes. Additionally, si‐hsa_circ_0042003 delivered by exosomes derived from U251 cells could reverse the resistance of glioma to TMZ, inhibiting cell proliferation and accelerating cell apoptosis, and restraining tumor growth in vivo. This study could provide ideas for the development of treatment strategies for patients with TMZ‐resistant glioma.
DISCLOSURE
The authors declare no conflict of interest.
ACKNOWLEDGMENTS
This study was supported by the Specific Research Fund for TCM Science and Technology of Guangdong Provincial Hospital of Chinese Medicine (YN2016QJ18) and the Special Project for Clinical Research of Guangdong Provincial Hospital of Chinese Medicine (YN10101902).
Si J, Li W, Li X, Cao L, Chen Z, Jiang Z. Heparanase confers temozolomide resistance by regulation of exosome secretion and circular RNA composition in glioma. Cancer Sci. 2021;112:3491–3506. 10.1111/cas.14984
Contributor Information
Jinchao Si, Email: sijinchao2003@163.com.
Zhi Jiang, Email: 2000jiangzhi@163.com.
REFERENCES
- 1.Ghiaseddin AP, Shin D, Melnick K, Tran DD. Tumor treating fields in the management of patients with malignant gliomas. Curr Treat Options Oncol. 2020;21(9):76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5‐year analysis of the EORTC‐NCIC trial. Lancet Oncol. 2009;10(5):459‐466. [DOI] [PubMed] [Google Scholar]
- 3.Muto J, Matsutani T, Matsuda R, Kinoshita M, Oikawa M, Pallud J. Temozolomide radiochemotherapy for high‐grade glioma patients with hemodialysis: a case series of 7 patients. Neuro‐Oncol Pract. 2020;7(1):111‐117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.López‐Valero I, Torres S, Salazar‐Roa M, et al. Optimization of a preclinical therapy of cannabinoids in combination with temozolomide against glioma. Biochem Pharmacol. 2018;157:275‐284. [DOI] [PubMed] [Google Scholar]
- 5.Iozzo RV, Sanderson RD. Proteoglycans in cancer biology, tumour microenvironment and angiogenesis. J Cell Mol Med. 2011;15(5):1013‐1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hammond E, Khurana A, Shridhar V, Dredge K. the role of Heparanase and sulfatases in the modification of Heparan sulfate proteoglycans within the tumor microenvironment and opportunities for novel cancer therapeutics. Front Oncol. 2014;4:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ilan N, Elkin M, Vlodavsky I. Regulation, function and clinical significance of Heparanase in cancer metastasis and angiogenesis. Int J Biochem Cell Biol. 2006;38(12):2018‐2039. [DOI] [PubMed] [Google Scholar]
- 8.Rivara S, Milazzo FM, Giannini G. Heparanase: a rainbow pharmacological target associated to multiple pathologies including rare diseases. Fut Med Chem. 2016;8(6):647‐680. [DOI] [PubMed] [Google Scholar]
- 9.Vlodavsky I, Singh P, Boyango I, et al. Heparanase: From basic research to therapeutic applications in cancer and inflammation. Drug Resist Updates. 2016;29:54‐75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin H, Cui M. New advances of heparanase and heparanase‐2 in human diseases. Arch Med Res. 2018;49(7):423‐429. [DOI] [PubMed] [Google Scholar]
- 11.Kundu S, Xiong A, Spyrou A, et al. Heparanase promotes glioma progression and is inversely correlated with patient survival. Mol Cancer Res. 2016;14(12):1243‐1253. [DOI] [PubMed] [Google Scholar]
- 12.Shteingauz A, Boyango I, Naroditsky I, et al. Heparanase enhances tumor growth and chemoresistance by promoting autophagy. Can Res. 2015;75(18):3946‐3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nazri HM, Imran M, Fischer R, et al. Characterization of exosomes in peritoneal fluid of endometriosis patients. Fertil Steril. 2020;113(2):364‐373.e362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daßler‐Plenker J, Küttner V, Egeblad M. Communication in tiny packages: exosomes as means of tumor‐stroma communication. Biochim Biophys Acta. 2020;1873(2):188340. [DOI] [PubMed] [Google Scholar]
- 15.Kulkarni B, Kirave P, Gondaliya P, et al. Exosomal miRNA in chemoresistance, immune evasion, metastasis and progression of cancer. Drug Discovery Today. 2019;24(10):2058‐2067. [DOI] [PubMed] [Google Scholar]
- 16.Milman N, Ginini L, Gil Z. Exosomes and their role in tumorigenesis and anticancer drug resistance. Drug Resist Updates. 2019;45:1‐12. [DOI] [PubMed] [Google Scholar]
- 17.Aghebati‐Maleki A, Nami S, Baghbanzadeh A, et al. Implications of exosomes as diagnostic and therapeutic strategies in cancer. J Cell Physiol. 2019;234(12):21694‐21706. [DOI] [PubMed] [Google Scholar]
- 18.Thompson CA, Purushothaman A, Ramani VC, Vlodavsky I, Sanderson RD. Heparanase regulates secretion, composition, and function of tumor cell‐derived exosomes. J Biol Chem. 2013;288(14):10093‐10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jia L, Tian Y, Chen Y, Zhang G. The silencing of LncRNA‐H19 decreases chemoresistance of human glioma cells to temozolomide by suppressing epithelial‐mesenchymal transition via the Wnt/β‐Catenin pathway. Onco Targets Ther. 2018;11:313‐321. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;30(1):1‐29. [DOI] [PubMed] [Google Scholar]
- 21.Ding C, Yi X, Wu X, et al. Exosome‐mediated transfer of circRNA CircNFIX enhances temozolomide resistance in glioma. Cancer Lett. 2020;479:1‐12. [DOI] [PubMed] [Google Scholar]
- 22.Wang X, Zhang H, Yang H, et al. Exosome‐delivered circRNA promotes glycolysis to induce chemoresistance through the miR‐122‐PKM2 axis in colorectal cancer. Mol Oncol. 2020;14(3):539‐555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo H, Chen Z, Wang S, et al. c‐Myc‐miR‐29c‐REV3L signalling pathway drives the acquisition of temozolomide resistance in glioblastoma. Brain. 2015;138(Pt 12):3654‐3672. [DOI] [PubMed] [Google Scholar]
- 24.Jabbari N, Akbariazar E, Feqhhi M, Rahbarghazi R. Breast cancer‐derived exosomes: tumor progression and therapeutic agents. J Cell Physiol. 2020;235(10):6345–6356. [DOI] [PubMed] [Google Scholar]
- 25.Asare‐Werehene M, Nakka K. The exosome‐mediated autocrine and paracrine actions of plasma gelsolin in ovarian cancer chemoresistance. Oncogene. 2020;39(7):1600‐1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hon KW, Ab‐Mutalib NS. Extracellular vesicle‐derived circular RNAs confers chemoresistance in colorectal cancer. Sci Rep. 2019;9(1):16497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramani VC, Zhan F, He J, et al. Targeting heparanase overcomes chemoresistance and diminishes relapse in myeloma. Oncotarget. 2016;7(2):1598‐1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borderud SP, Li Y, Burkhalter JE, Sheffer CE, Ostroff JS. Electronic cigarette use among patients with cancer: characteristics of electronic cigarette users and their smoking cessation outcomes. Cancer. 2015;121(5):800. [DOI] [PubMed] [Google Scholar]
- 29.Fu X, Liu M, Qu S, et al. Exosomal microRNA‐32‐5p induces multidrug resistance in hepatocellular carcinoma via the PI3K/Akt pathway. J Exp Clin Cancer Res. 2018;37(1):52. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Wang M, Qiu R, Yu S, et al. Paclitaxel‐resistant gastric cancer MGC‐803 cells promote epithelial‐to‐mesenchymal transition and chemoresistance in paclitaxel‐sensitive cells via exosomal delivery of miR‐155‐5p. Int J Oncol. 2019;54(1):326‐338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang B, Zhang Y, Ye M, Wu J, Ma L, Chen H. Cisplatin‐resistant MDA‐MB‐231 Cell‐derived exosomes increase the resistance of recipient cells in an exosomal miR‐423‐5p‐dependent manner. Curr Drug Metab. 2019;20(10):804‐814. [DOI] [PubMed] [Google Scholar]
- 32.Sharma P, Mesci P, Carromeu C, et al. Exosomes regulate neurogenesis and circuit assembly. Proc Natl Acad Sci USA. 2019;116(32):16086‐16094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang W, Wang J. Exosomes and their noncoding RNA cargo are emerging as new modulators for diabetes mellitus. Cells. 2019;8(8):853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoshino A, Costa‐Silva B, Shen TL, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329‐335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pereira AL, Magalhães L, Pantoja RP, Araújo G, Ribeiro‐Dos‐Santos Â, Vidal AF. The biological role of sponge circular RNAs in gastric cancer: main players or coadjuvants? Cancers. 2020;12(7):1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jeyaraman S, Hanif EAM, Ab Mutalib NS, Jamal R, Abu N. Circular RNAs: potential regulators of treatment resistance in human cancers. Front Genet. 2019;10:1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang M, Yu F, Li P, Wang K. Emerging function and clinical significance of exosomal circRNAs in cancer. Mol Ther Nucleic Acids. 2020;21:367‐383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vlodavsky I, Gross‐Cohen M, Weissmann M, Ilan N, Sanderson RD. Opposing functions of heparanase‐1 and heparanase‐2 in cancer progression. Trends Biochem Sci. 2018;43(1):18‐31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okada H, Hirano D, Taniguchi T. Negative symptoms in schizophrenia: modeling the role of experience factor and expression factor. Asian J Psychiatry. 2020;53:102182. [DOI] [PubMed] [Google Scholar]
- 40.Yan L, Liu G, Cao H, Zhang H, Shao F. Hsa_circ_0035483 sponges hsa‐miR‐335 to promote the gemcitabine‐resistance of human renal cancer cells by autophagy regulation. Biochem Biophys Res Comm. 2019;519(1):172‐178. [DOI] [PubMed] [Google Scholar]
- 41.Hong W, Xue M, Jiang J, Zhang Y, Gao X. Circular RNA circ‐CPA4/let‐7 miRNA/PD‐L1 axis regulates cell growth, stemness, drug resistance and immune evasion in non‐small cell lung cancer (NSCLC). J Exp Clin Cancer Res. 2020;39(1):1‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]


