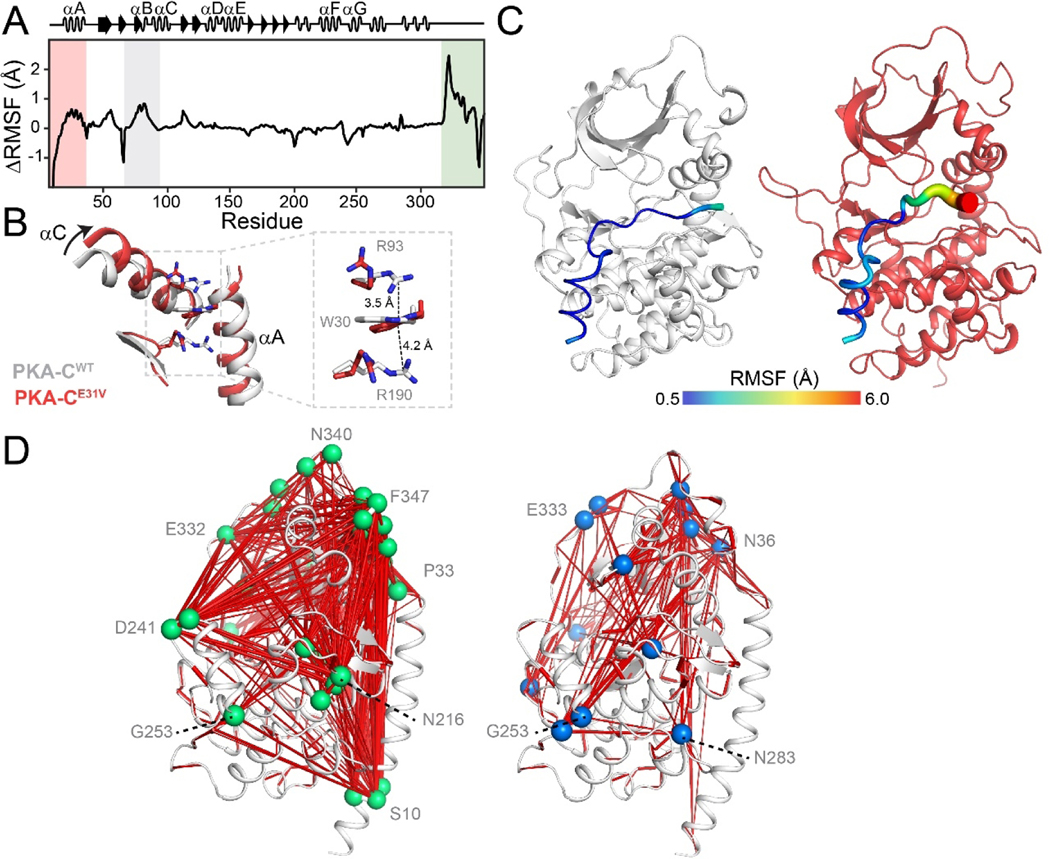
Figure 5. Altered conformational dynamics of PKA-CE31V revealed by MD simulations.
(A) ΔRMSF of PKA-CE31V in the binary form over 1.0 μs of simulation. (B) Overlay of PKA-CWT (PDB: 1ATP; gray) and PKA-CE31V (from MD simulations; red) highlighting the structural rearrangements of the αA- and αC-helices caused by the E31V mutation. The inset shows the cation-π stacking interactions altered in response to the mutation. (C) Distinct dynamics of the pseudosubstrate, PKI5–24, in the ternary complex. Putty representation of RMSF for PKI5–24 for a typical MD snapshot of PKA-CWT (gray) and PKA-CE31V (red). (D)Comparison of allosteric mapping from mutual information matrices (MIcutoff = 0.3), with the key hub residues labeled for PKA-CWT (right) and PKA-CE31V (left).