Abstract
Hydrogels have been widely investigated in biomedical fields due to their similar physical and biochemical properties to the extracellular matrix (ECM). Collagen and hyaluronic acid (HA) are the main components of the ECM in many tissues. As a result, hydrogels prepared from collagen and HA hold inherent advantages in mimicking the structure and function of the native ECM. Numerous studies have focused on the development of collagen and HA hydrogels and their biomedical applications. In this extensive review, we provide a summary and analysis of the sources, features, and modifications of collagen and HA. Specifically, we highlight the fabrication, properties, and potential biomedical applications as well as promising commercialization of hydrogels based on these two natural polymers.
Keywords: Microstructure, Fabrication, semi-interpenetrating network, dynamic properties, extracellular matrix
1. Introduction
Hydrogels are crosslinked networks of hydrophilic polymers with high water content that have been studied and used clinically for many decades. They have been used extensively to investigate the interactions of cells with their microenvironment and as scaffolds for biomedical and tissue engineering applications, such as drug delivery and wound dressings. Among the various polymers available, those derived from natural biological sources add significant bioactivity and thus are widely used [1–3].
Native ECMs are mainly composed of proteins, glycosaminoglycans (GAGs), which are polysaccharides that often bind covalently to a protein backbone to form proteoglycans [4]. Collagen is the most abundant protein in the ECM and in mammals [5]. Cells interact with collagen via various cell surface receptors such as integrins [6]. Collagen can be degraded by matrix metalloproteinases, which play an important role in the remodeling of the ECM and the development of tissues [7]. Collagen can be sourced from a variety of animals and tissues and is thus widely available for research and clinical applications. To overcome batch-to-batch variability and possible immunogenicity of animal-derived collagen, recombinant collagen has been explored as another source to mimic human collagen [8,9]. Collagen-based hydrogels have been fabricated through the self-assembly of collagen fibrils or by adding chemical crosslinking reagents. However, potential modifications of collagen are limited by both the complexity of its structure and biocompatibility requirements. Commercial applications of collagen matrices include wound repair, skin healing, and orthopedic regeneration (see Section 3.3) [10].
HA is a highly hydrated GAG distributed in the ECMs of various types of tissues and is important for diverse biological processes and tissue functions (see Section 2.2). The different functional groups in HA contain carboxyl, hydroxyl, and acetyl groups, which enable chemical modifications [11] that can alter the properties of the resulting material. HA-based hydrogels are biocompatible, have tunable properties, and mainly interact with CD44 membrane receptors found on many cell types. Consequently, the design, fabrication, and biomedical application of HA-based hydrogels have been widely pursued in recent years [12].
Hydrogels containing collagen and HA combine the features of these two ECM components to mimic the ECM in both structure and function. In physically crosslinked collagen and HA blend (ColHA) hydrogels, collagen molecules aggregate due to electrostatic and hydrophobic interactions to form fibrils, whereas HA can associate with the surface of collagen fibrils or occupy interstitial space and influence the microstructure and viscosity of the hydrogel (see Section 5.1.2) [13–15]. Chemical crosslinking can be used to tailor the properties of ColHA hydrogels (e.g., mechanical properties and microstructure) for applications in vitro and in vivo. In addition to advances in crosslinking chemistry, numerous engineering techniques, such as plastic compression, molding, and bioprinting, have been developed to fabricate collagen- and HA-based hydrogels with tunable stiffness and spatially-defined microstructure [16,17]. The combination of stimuli-responsive chemistries and biofabrication technologies can be used to tailor hydrogel degradation, mechanical strength, and structural patterning to advance applications such as guiding cellular behavior and disease therapy.
The objective of this review is to provide a comprehensive understanding of collagen and HA biopolymers and their use within both individual hydrogels and combined ColHA hydrogels. Although collagen and HA have been reviewed extensively, other review papers have not given much attention to hydrogels that combine both components. In this review, the sources, structures, bioactivities, production, and applications of HA and collagen polymers are summarized (Sections 2 and 3). Current crosslinking and fabrication strategies of collagen, HA, and ColHA hydrogels are discussed in greater detail (Section 4). The regulation of internal fibril and porous microstructure as well as the mechanical properties and dynamic control of ColHA hydrogel properties are discussed (Section 5). Recent applications of collagen- and HA-based hydrogels in tissue engineering, including bone and cartilage regeneration, skin repair, and disease models are described (Section 6). Finally, the future directions of collagen and HA based hydrogels, ranging from control of hydrogel properties to biomedical applications, are considered (Section 7).
2. In vivo roles and functions of collagen and HA
The ECM is an acellular network of macromolecules present within tissues that provides structural support and biomechanical and biochemical cues to the surrounding cells. The ECM is insoluble, which is mainly due to highly cross-linked ECM proteins such as collagens [18], and thus is a stable source of cell signaling molecules. Despite the variation in composition between the ECM of different tissues, all ECMs are composed of three main classes of molecules: proteins, glycoproteins, and proteoglycans. The most abundant fibrous protein found in the mammalian ECM is collagen, which comprises ≤30% of total body protein mass [19]. Proteoglycans are biomacromolecules in which proteins are covalently attached to GAGs. They offer unique buffering, hydrating, and force-resisting properties to the ECM [19]. HA is a unique GAG as it does not contain sulfate, and it does not contain covalently bound core proteins [20]. Instead, HA non-covalently binds to aggrecans though link protein [21,22]. The high viscosity of HA confers resistance to compressive forces and makes it especially important in load-bearing tissues. HA cell receptors regulate functions including induction of chondrogenesis, osteogenesis, neurogenesis, cardiogenesis, and angiogenesis; increasing proliferation of cells including astrocytes and endothelial cells; and controlling inflammation by binding monocytes [23]. In the native environment, collagen and HA create semi-interpenetrating networks [24]. This review will specifically focus on collagen and HA. For a detailed review of glycoproteins, see the 2020 review by Walimbe and Panitch [25].
2.1. Distribution and function of collagen and HA in tissues
2.1.1. Collagen types, properties, and distribution
Collagen refers to a family of proteins defined by their unique structural motif. Each collagen monomer is composed of three polypeptide chains (referred to as α-chains [26]) that self-assemble in a zipper-like fashion to form a collagen monomer, which in turn assembles further into a higher-order supramolecular right-handed triple-helical domain [27,28]. To date, 28 collagen types have been identified. They are designated with Roman numerals I to XXVIII in order of their discovery and are composed of different combinations of at least 46 types of polypeptide chains [27,29]. Collagen monomers composed of the same three polypeptide chains are referred to as a homotrimer, whereas collagen monomers with different polypeptide chains are heterotrimers. The polypeptide chains are composed of a repeated tripeptide motif (GXY)n. Glycine (G), which is sterically small, occurs at every third residue and allows for close packing of the polypeptide chains to form a triple helix about a common axis [6]. The identities of the X and Y residues are dependent on the collagen type; however, Y is often the post-translationally modified residue 4-hydroxyproline [30]. Triple-helical collagenous (COL) domains are interrupted by non-collagenous (NC) domains that confer different biological activities depending on the collagen type [6,31]. The convention for domains typically begins with numbering from the C-terminus of collagen [6]. The number and locations of the NC domains contribute to different supramolecular organization, which determines the function of the collagens [6,32]. Collagens are classified into different subfamilies based on their structure and function. The subfamilies include fibrillar collagens (types I, II, III, V, XI, XXIV, and XXVII), fibril associated collagens with interrupted triple helices (FACIT) and related collagens (types IX, XII, XIV, XVI, XIX, XX, XXI, and XXII), beaded filament-forming collagen (type VI), basement membrane and associated collagens (types IV, VII, XV, and XVIII), transmembrane collagens (types XIII, XVII, XXIII, and XXV), and hexagonal network collagens (types VIII and X) [6,33,34]. Collagen types and their defining characteristics are summarized in Table 1. As fibrillar collagens are the most abundant ECM proteins, they are on the forefront of tissue engineering research and thus will be the focus of this review.
Table 1.
Defining features of collagen types.
| Collagen Type | Subfamily | Molecular Species | Supramolecular Structure and Structural Features | Distributions | Function |
|---|---|---|---|---|---|
| I | Fibril Forming | • [α1(I)]2, α2(I) • [α1(I)]3 |
300 nm molecule, 67 nm banded fibril [35] | Ubiquitous, predominant in skin, tendons, ligaments, bones [35] | Key structural component of tissues [35] |
| II | Fibril Forming | • [α1(II)]3 | Cartilage | Confers tensile strength and elasticity to cartilage [36], endochondral bone formation [37] | |
| III | Fibril Forming | • [α1(III)]3 | Stabilizing C-terminal cystine knot [38] | Dermis, aorta [28], uterus, blood vessels, bowel, wound healing, blood clotting cascade [39] | Tensile strength and integrity [39] |
| IV | basement membrane and associated collagens | • [α1(IV)]2, α2(IV) • α3(IV), α4(IV), α5(IV) • [α5(IV)]2, α6(IV) |
COL domain with 21–26 interruptions [40] | Basement membranes | Cell adhesion, migration, differentiation, growth [41] |
| V | Fibril Forming | • [α1(V)]2, α2(V) • [α1(V)]3 • [α1(V)]2, α4(V) • α3(XI), α1(V), α3(XI) |
Thrombospondin domain [6] | Interstitial tissue [42], dermal-epidermal junction [43] | Regulates collagen fibrillogenesis [44] |
| VI | beaded filamentforming collagen | • α1(VI),α2(VI),α3(VI) | C-terminal propeptide endotrophin hormone [45,46] Much lower frequency of GPO repeat [28] |
Ubiquitous [28], basement membrane-interstitial matrix interface [46] | Maintains the integrity of skeletal muscle [46] |
| VII | basement membrane and associated collagens | • [α1(VII)]3 | Exceptionally long triplehelix domain, Kunitz domain [6] | Epidermal–dermal Junction [6] | Anchoring collagen, binds fibril forming collagens [47] |
| VIII | hexagonal network collagens | • [α1(VIII)]2, α2(VIII) • α1(VIII), [α2(VIII)]2 • [α1(VIII)]3 • [α2(VIII)]3 |
C1q domain [6] | Descemet’s membrane [48], vascular smooth muscle [49] | Mechanical stability of vascular wall, bridge between ECM components [50] |
| IX | FACIT | • α1(IX), α2(IX), α3(IX) | Thrombospondin domain, three COL and three NC domains [6] | Articular cartilage [51] | Stabilizes fibrillar collagen network in the cartilage matrix, anchors matrilin 3 and proteoglycans [52] |
| X | hexagonal network collagens | • [α1(X)]3 | C1q domain [6] | Hypertrophic cartilage [53] | Regulates endochondral ossification of articular cartilage [54,55] |
| XI | Fibril Forming | • α1(XI), α2(XI), α3(XI) • α1(XI), α1(V), α3(XI) |
Thrombospondin domain [56] | Minor component of hyaline cartilage collagen fibrils [57], broadly distributed in testis, trachea, tendons, trabecular bone, skeletal muscle, placenta, lung, and the neoepithelium of the brain [56] | Nucleates and controls cartilage collagen fibril formation [57] |
| XII | FACIT | • [α1(XII)]3 | Largest FACIT collagen, Triple armed NC3 domain [58], two variants [59] NC3 domain carries glycosaminoglycan chains [59] | Mesenchymal tissue during development [60], periodontal ligament [61], dermis around hair follicles [62], cornea of the eye [63] in adults | Temporarily stabilizes collagen fibrils during development [59,60,64] |
| XIII | transmembrane collagens | • [α1(XIII)]3 | Connective tissue, blood vessel and junctions [65] | Bone formation [66], regulates formation of neuromuscular synapse [67] | |
| XIV | FACIT | • [α1(XIV)]3 | Skin, tendon, cornea, articular cartilage [68] | Fibrillogenesis regulation, Maintaining mechanical integrity [68], embryonic development [60] | |
| XV | basement membrane and associated collagens | • [α1(V)]3 | Bonded to chondroitin sulfate via disulfide-bonds [69] Flexible due to knot/figure-of-eight/pretzel configuration [70] Thrombospondin domain [6] |
Cardiac and skeletal muscles, basement membrane zones [71] | Maintains integrity of ECM [71] |
| XVI | FACIT | • [α1(VI)]3 | Flexible due to kinks in structure, 10 COL and 11 NC domains [72–74] | Territorial cartilage matrix [74,75], integrated into fibrillin-1-rich microfibrils containing in skin [74] | Hypothesized to stabilize ECM by organizing and connecting fibrillar networks, cell adhesion and invasion [72,76] |
| XVII | transmembrane collagens | • [α1(XVII)]3 | 15 COL and 16 NC domains [6] | Basement membrane zone, specifically hemidesmosomes [77,78], central nervous system neurons [79] | Epidermal cell adhesion [80] Proliferation of epidermis [81] |
| XVIII | basement membrane and associated collagens | • [α1(XVIII)]3 | Thrombospondin domain, 10 triple helical COL domains 11 NC domains [6] | Basement membrane zones [82] | Eye development [83], maintaining basement membrane integrity [84] |
| XIX | FACIT | • [α1(XIX)]3 | Thrombospondin domain [6] | Vascular, neuronal, mesenchymal, epithelial basement membrane zones [85], hippocampal neurons [86] | Affects the phenotype for smooth muscle motor dysfunction and hypertension sphincter [87] |
| XX | FACIT | • [α1(XX)]3 | von Willebrand factor A, fibronectin type III repeat, thrombospondin domain [6,88] | Possibly bile ducts, breast, cerebellum, smooth muscle cells [89] | |
| XXI | FACIT | • [α1(XXI)]3 | two collagenous domains interrupted by three noncollagenous domains [6,90] | heart, stomach, kidney, skeletal muscle, placenta [90] | May play a role in blood vessel assembly [91] |
| XXII | FACIT | • [α1(XXII)]3 | Does not directly polymerize with fibrillar collagens, but rather associates with components of microfibrils such as fibrillins [92] | Tissue junctions: myotendinous junctions, articular cartilage-synovial fluid junction, border between the anagen hair follicle and the dermis in the skin [92] | Possibly mechanical stability of myotendinous junctions [92] |
| XXIII | transmembrane collagens | • [α1(XXIII)]3 | N-terminal cytoplasmic domain, a transmembrane domain, and extracellular triple helical domains [93] | Lung, cornea, skin, tendon, amnion [94] | Cancer, cell-cell and cell-matrix adhesion mediation [95] Expression elevated in prostate cancer recurrence and distant metastases [96] |
| XXIV | FACIT | • [α1(XXIV)]3 | Thrombospondin-Nterminal like motif and charged segments with tyrosine residues on amino-terminal domain [97] | Bone and cornea [97] | Marker of osteoblast differentiation and bone formation [98] |
| XXV | transmembrane collagens | • [α1(XXV)]3 | COL domain interrupted four times (two four-residue imperfections and two large NC sequences) [99] | Neurons predominantly of brain, also of heart, testis, eye [100–102] | Also known as collagen-like amyloidogenic component, isolated from Alzheimerdiseases brains, component of amyloid plaques [99] |
| XXVI | Not assigned | • [α1(XXVI)]3 | Two collagenous regions and no obvious sequence homology [103] | Testes, ovary [103] | Testis and ovary development [103] |
| XXVII | Fibril Forming | • [α1(XXVII)]3 | Nonstriated fibrils, 10 to 80 nm fibril width [104,105] | Adult cartilage [106] | Cartilage calcification, possibly cartilage transition to bone [107] |
| XXVIII | Not assigned | • [α1(XXVIII)]3 | 528 amino-acid collagenous domain flanked by two von Willebrand factor A [6,108] Structurally resembles collagen VI |
Basement membranes of peripheral nervous system [109,110] |
2.1.1.1. Fibrillar collagens
Fibrillar collagens are ubiquitous throughout the ECM space of tissues and are most abundant in bones, blood vessels, skin, tendons, and fibrous capsules of organs [111,112]. The ability of the protein to self-assemble into fibers confers mechanical strength and provides a structural framework for tissues and organs. The formation of fibrillar collagens in vivo is reviewed by Kadler [113]. Briefly, cells secrete thin collagen fibrils into the ECM via fibripositor projections from the plasma membrane. Fibrillar collagen assembly is heavily regulated in the body by cellular and tissue-specific stimuli and results in widely varying collagen fibril architectures from aligned bundles in tendons to woven matrices in skin.
The fibrillar collagen family can be further subdivided into the major fibril-forming collagens, which consist of type I, II, and III, and minor fibril-forming collagens, which are composed of types V and XI [114]. Major fibrillar collagens are the main structural components of the ECM and are supplemented by the minor fibrillar collagens. The amino acid sequences of the α-chains of minor fibrillar collagens share a high degree of similarity with major fibrillar collagens with the exception of variable region domains caused by alternative splicing of the proteins [114]. The function of minor fibrillar collagens is not yet fully understood; however, the manifestation of diseases associated with their mutations highlight their importance. For an in-depth review on minor fibrillar collagens, refer to the 2012 review paper by Fang and coworkers [114].
In fibrillar collagen, the X position of the GXY motif is commonly proline (P). The Y position of the GXY motif is typically 4-hydroxyproline (O), which is post-translationally modified proline (P) [115], or lysine, which is post-translationally modified to hydroxylysine [116]. In human collagen, proline and lysine are 42–54% and 13–28% hydroxylated, respectively [117,118]. Hydroxyproline is an imino acid predominantly found in collagens, which contain 99.8% of the body’s hydroxyproline content [119]. The GPO triplet is the most frequently observed amino acid sequence in collagen and comprises ~10.5% of the collagen content [120]. Each of the amino acids in the triplet contributes to the supramolecular structure. Glycine enables a packed structure of the collagen monomer chains, which is stabilized by intramolecular hydrogen bonding and electron-withdrawing effects of hydroxyproline [115]. Proline provides local conformational flexibility, which enables low energy molecular compression, extension, and bending [121]. The hydroxyl groups of 4-hydroxyproline stabilize the collagen triple helix. Nearly complete hydroxylation of the prolines in the Y position is necessary for the formation of collagen that is stable at 37 °C [30]. The Brodsky group elucidated this stabilizing effect of the triple helix by varying the amino acids in the guest X and Y positions in collagen mimetic peptides (CMPs) containing GXY repeats (Table 2) [122]. The sterically hindered proline and hydroxyproline imino acids in collagen, combined with the small glycine residue, result in bending and twisting of individual monomer chains to form left-handed polyproline-II helices [27]. Adzhubei and colleagues provide an in-depth review on proline-II structure, properties, and physics [123]. Three polyproline-II helices self-assemble into a right-handed helix that comprises the procollagen molecule (Fig. 1a). The importance of charged amino acids in triple helix formation is summarized in a review by the Kiick group [124]. CMPs and collagen-like proteins from bacteria have been used to understand collagen triple helix formation and how to meet or exceed helical thermal stability [124,125] by replacing hydroxyproline with non-canonical amino acids such as fluoroproline and chloroproline [126–128], incorporating electrostatic interactions of oppositely-charged residues either within or between a tri-peptide chain repeat [129–137], and using numerous other methods [9].
Table 2.
a Amino acids impart varying levels of thermal stability, measured as melting temperature (Tm), in the X and Y guest residue positions of the CMP triple helix.
| Guest Residue | Gly-X-Hyp Tm (°C) | Gly-Pro-Y Tm (°C) |
|---|---|---|
| Pro | 47.3 | - |
| Hyp | - | 47.3 |
| Glu | 42.9 | 39.7 |
| Ala | 41.7 | 40.9 |
| Lys | 41.5 | 36.8 |
| Arg | 40.6 | 47.2 |
| Gln | 40.4 | 41.3 |
| Asp | 40.1 | 34.0 |
| Leu | 39.0 | 31.7 |
| Val | 38.9 | 40.0 |
| Met | 38.6 | 42.6 |
| Ile | 38.4 | 41.5 |
| Asn | 38.3 | 30.3 |
| Ser | 38.0 | 35.0 |
| His | 36.5 | 35.7 |
| Thr | 36.2 | 39.7 |
| Cys | 36.1 | 37.7 |
| Tyr | 34.3 | 30.2 |
| Phe | 33.5 | 28.3 |
| Gly | 33.2 | 32.7 |
| Trp | 31.9 | 26.1 |
Adapted with permission from A.V. Persikov, J.A.M. Ramshaw, A. Kirkpatrick, B. Brodsky, Biochemistry 39 (2000) 14960–14967 [122]. Copyright 2000 American Chemical Society.
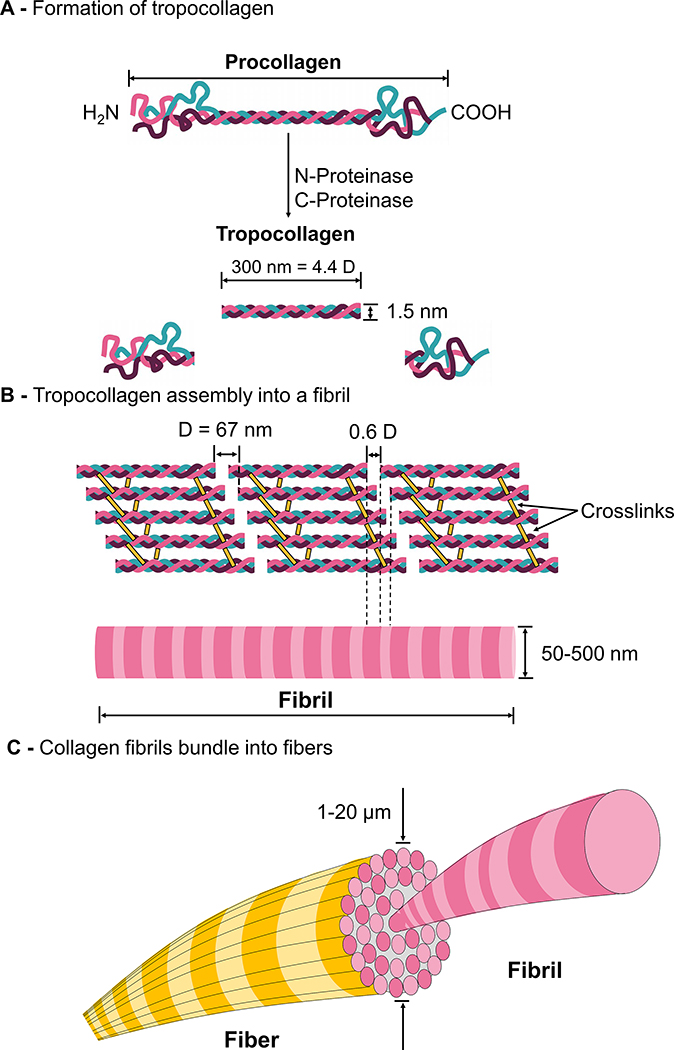
Fig. 1.
Molecular structure and synthesis of collagen I. (A) Following intracellular post translational modifications, three polypeptide chains assemble into procollagen, which is then exocytosed into the extracellular space. N-proteinase and C-proteinase cleave the ends of procollagen to form tropocollagen. (B) Collagen I tropocollagen is 300 nm (corresponding to 4.4 D) in length and 1.5 nm in diameter. Tropocollagen molecules self-organize and crosslink to form collagen fibril with periodicity of D, which is typically 64–67 nm. (C) Collagen fibrils bundle to form fibers.
Despite the recurrence of the stabilizing GPO amino acid sequence, irregularities in the X and Y residues are essential for biological specificity and function [130]. Hydroxylysine serves as a substrate for O-linked glycosylation, which allows for crosslinking of collagen [33]. The degree of glycosylation affects the packaging of mature molecules into fibrils, and increased glycosylation decreases fibril diameter.
The biosynthesis pathway of fibrillar collagen has been studied in great detail and has been extensively reviewed by Bornstein in 1974 [138]. Briefly, fibrillar collagen synthesis begins with synthesis of procollagen precursors that contain NC regions on either end. After post-translational modifications, including the formation of hydroxylysine, 4-hydroxyproline, and the glycosylation of hydroxylysine [116], disulfide bonding occurs between polypeptide chains that compose the monomer. Next, the proteins begin to assemble from the C-terminus by twisting tightly towards the left and propagating toward the N-terminus to form procollagen (Fig. 1A). The procollagen molecules are secreted into the ECM [33] upon which the globular N- and C-termini are cleaved off to form tropocollagen. The tropocollagen is approximately 300 nm long and has a diameter of 1.5 nm [35].
On the macromolecular level, five tropocollagen molecules arrange head-to-tail into parallel structures directed by electrostatic and hydrophobic interactions and form a microfibril, the basic subunit of a collagen fibril (Fig. 1B) [35,139]. During this process, lysine and hydroxylysine undergo oxidative deamination into reactive aldehydes that participate in spontaneous crosslink formation between the tropocollagens, and the result are collagen fibrils [30,140]. The distance between two staggered tropocollagen molecules is 64–67 nm (Fig. 1B) [35]. This distance comprises the overlap (distance tropocollagens are staggered by) and gap (distance between two adjacent tropocollagen molecules). This measurement of 64–67 nm is termed distance D, and other dimensions on the collagen molecule are listed in terms of D [35]. The tropocollagen molecules are 4.4 D in length, and the gap between the ends of two adjacent nonoverlapping tropocollagen molecules is 0.6 D. Thus, this regular parallel structure with gaps confers fibrous collagens their signature banded appearance that can be observed via electron microscopy. The gap appears as the dark region of the negatively stained TEM images of collagen fibrils, whereas the overlap region appears as a lighter band due to inability of the stain to penetrate into the region [113,141]. These microfibrils further associate to form fibrils with diameters that range from 50 to 500 nm depending on the tissue [13]. Fibrils bundle into fibers that are 1–20 μm in diameter (Fig. 1C) [142].
2.1.1.2. Most abundant fibrillar collagens
Collagen I is the most abundant fibrillar collagen and makes up 70% of total collagen [112]. It is primarily found in skin, ligaments, bones, and tendons but is ubiquitous throughout the body. Collagen I has two structural variations. The most abundant variant consists of two identical polyproline-II chains α1(I) and one α2(I) chain (heterotrimeric). A smaller portion of collagen I consists of homotrimers with three α1(I) chains [32]. Collagen V is a minor fibrillar collagen often associated with collagen I [28]. For a detailed review on collagen I, including splicing, regulation, and transcriptional regulation, see the 2002 review by Rossert and Brombrugghe [35].
Collagen II is the major constituent of hyaline cartilage, where it constitutes approximately 90% of the total collagen [143]. It is also the major collagen of vitreous humor and nucleus pulposus of intervertebral discs [34]. Other locations include the tendon, retina, sclera, the lens of the eye, notochord, heart, and brain [28,144,145]. Collagen II is a homotrimeric molecule composed of [α1(II)]3 chains [6]. Nearly half of the hydroxylysine residues on collagen II are glycosylated, compared to only 2 residues per chain in collagen I [28,146]. The glycosylation is hypothesized to affect fibril regulation and lateral growth [28]. Collagen XI is a minor fibril forming collagen associated with collagen II. Collagen XI regulates fibrillogenesis by maintaining the spacing and diameter of collagen II fibrils [147].
Collagen III constitutes more than half of the total collagen and >20% of adult skin [148]. It is a homotrimer that is synthesized from three α1(III) chains [6]. Collagen III accompanies collagen I in almost every tissue in different ratios [28]. The protein confers structural integrity to hollow organs that must withstand stretching, including arteries, uterus, and bowel [39].
2.1.1.3. Other Collagen Sub-Families
FACIT are a family of collagens that do not themselves form fibrils but instead bind in regular intervals to the surface of fibril-forming collagens [6]. They are characterized by the presence of NC domain interruptions in the COL domain and the cystine-containing motif GXCXXXC [149]. By incorporating into the interfibrillar space of fiber-forming collagens, FACIT collagens alter surface properties and fibril assembly. They are believed to be involved in fibril-fibril interactions, as well as fibril interactions to other macromolecules present in the ECM.
Other types and families of collagens are less understood and an area of active research. Table 1 summarizes key features of the aforementioned collagens along with other collagen types not discussed here. For an in-depth discussion of other types of collagens, see the 2010 review by Bachinger et al. [28].
2.1.1.4. Collagen Degradation
Collagen is broken down by matrix metalloproteinases (MMPs) into small soluble peptides and amino acids. MMPs are zinc-dependent endopeptidases and typically require calcium as a cofactor [6,7]. MMPs were originally named for the substrates they were observed to degrade [150]. For example, MMP1, MMP3, and MMP8 are known as collagenases, and MMP2 and MMP9 are known as gelatinases. However, MMPs are capable of degrading more than their corresponding substrate [150]. Fibrillar collagens (I, II, and III) are degraded by MMP1, MMP2, MMP8, MMP13, and MMP14 [6,151]. MMP1 and MMP8 favor collagen I and collagen III, whereas MMP13 preferentially cleaves collagen II. Denatured collagens and collagen IV are degraded by MMP2 and MMP9. Collagens are broken down starting from the exterior of triple helices by these MMPs [7]. Collagen fragments are further degraded by gelatinases and nonspecific proteases [7,151]. Under physiological conditions, collagen in its triple helical form is largely not degraded by common proteases such as pepsin, trypsin, and papain [7,152].
2.1.2. HA properties and distribution
HA is an anionic, linear GAG primarily found in the ECM of soft connective tissues. Unlike other GAGs, HA is non-sulfated and does not covalently bind to a core protein to form a proteoglycan [153]. HA forms non-covalent bonds with aggrecan, and these bonds are stabilized by the link protein [21,22]. The molecular weight (MW) of HA in the human body typically ranges from 10 [154] to 8000 kDa [155], whereas other GAGs are typically between 15 to 20 kDa [153]. The different MW of HA confers various biological and biomechanical functions.
The term “hyaluronic acid” was created by Meyer and Palmer, who successfully extracted the compound from cattle vitreous humor, and subsequently named it hyaloid (meaning vitreous) and uronic acid (Fig. 2A), one of the sugar molecules that constitutes the polymer [156]. The term hyaluronan was introduced to conform with international polysaccharide nomenclature [157]. The polyanion without its corresponding cation is referred to as hyaluronate. Under physiological conditions, HA occurs in the salt form [158,159].
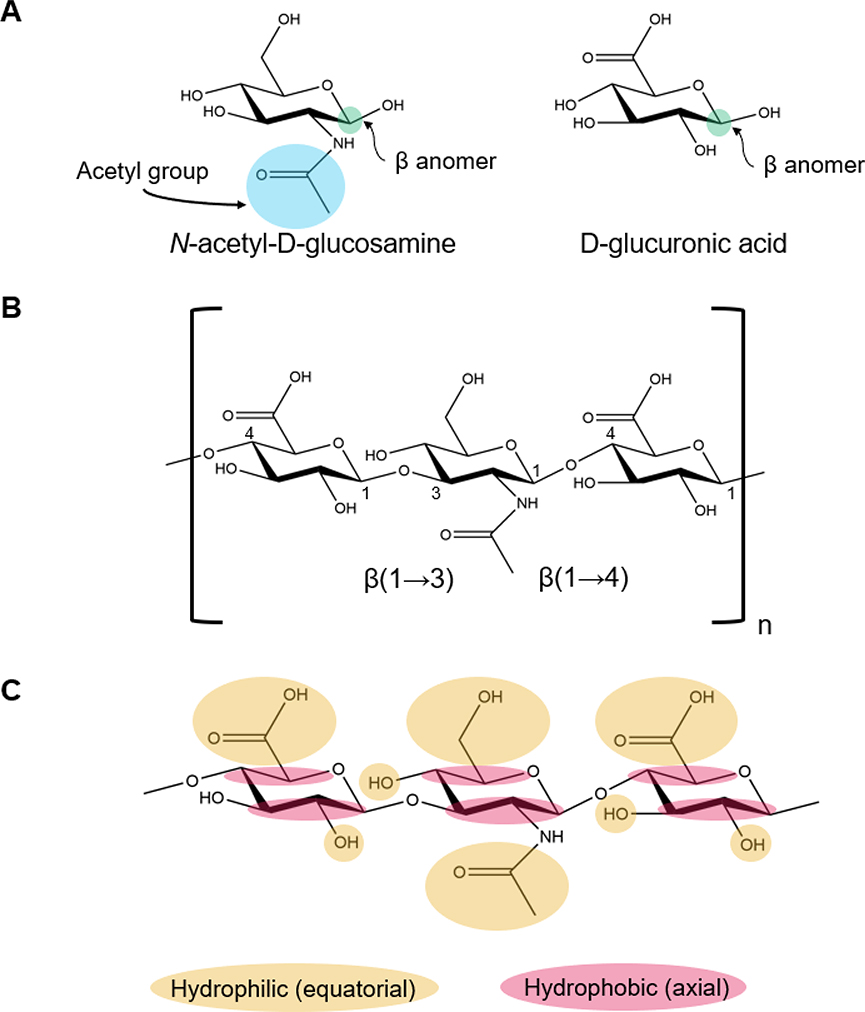
Fig. 2.
Chemical structure of HA and its components. (A) Structures of D-glucuronic acid and N-acetyl-D-glucosamine. (B) Chemical structure of the repeating disaccharide unit of HA linked together by alternating β-(1→4) and β-(1→3) glycosidic bonds. (C) Hydrophilic and hydrophobic moieties, which drive the twisting of the HA molecule, are labeled to show their equatorial and axial positions, respectively.
2.1.2.1. Chemical Structure and Conformation
HA is composed of repeating disaccharide units of D-glucuronic acid (GlcUA) and N-acetyl-D-glucosamine (GlcNAc) (Fig. 2A), which are linked together by alternating β-(1→4) and β-(1→3) glycosidic bonds (Fig. 2B) [160]. The resultant HA polymer is stable because bulky groups are in sterically favorable equatorial positions (Fig. 2C) [158]. The chemical structure of HA is identical across vertebrates and a few pathogenic bacteria that produce it [161,162].
Despite the seeming simplicity of the structure, assessment of the conformation of the molecule and the driving forces behind its molecular conformation is challenging. The natural rigidity of the polysaccharide chain is derived from restrictions on the rotation of the glycosidic bonds due to the bulky N-acetyl groups adjacent to the glycosidic bonds [159]. The conformation of the molecule is highly dependent on the counterion type, pH, temperature, extent of hydration, and, in the case of HA solutions, its concentration [160].
The arrangement of the hydrophobic axial hydrogens and hydrophilic polar equatorial groups drive twisting of the HA structure in solution (Fig. 2C) [158]. The repeating disaccharide provides regularity in spacing and makes twisting into a helical structure possible [159]. The backbone of HA in a physiological solution stiffens due to hydrogen bonding between the hydroxyl groups along the chain and interactions with the solvent [158]. This rigidity results in an extended random-coil configuration [153]. The coils in solution entangle at HA concentrations of 0.5–1 mg/mL and above and thus result in a substantial increase in viscosity [163]. The entangled HA networks create steric exclusions that reduce the mobility of HA and restrict the diffusion and hydrodynamic transport of other substances through the ECM [153]. For a detailed review on HA structure in solution and its physics, refer to reference [160].
2.1.2.2. Properties and Functions
Distinctive viscoelastic properties of hydrated HA confer its unique functionality. The HA polymer has an extraordinary ability to retain up to 1000 times its weight in water [164]. The high water absorption rate confers high viscosity to HA solutions even at lower concentrations [165]. Viscosity and elasticity of the hydrated polymer vary with shear rate, where high shear rate decreases viscosity and increases elasticity, which is the ability to store energy and facilitate recovery from the deformation [153]. The viscoelastic behavior is dependent on the solution conditions, notably the pH, which drastically changes the viscosity of the solution based on the state of entanglement, bonding, and electrostatics [166].
Depending on the MW, HA can influence cell proliferation, migration, morphogenesis, tissue inflammation, tumor development, and tumor metastasis among others [158,167]. HA can be classified into low (10–250 kDa), medium (250–1000 kDa), and high (>1000 kDa) MW HA [168]. High MW HA is found in most tissues and is involved in maintaining the integrity of the ECM, homeostasis, wound healing [169], anti-inflammatory response [170], and cell growth [171]. Low MW HA activates the proinflammatory [172] and macrophage response [173]. It also induces lymphangiogenesis [174] and angiogenesis [175].
2.1.2.3. Distribution in Tissues
HA constitutes about 15 grams of a 70 kg individual [167], and approximately one third of all HA turns over daily [176]. It is present in virtually all human tissues with the highest concentration of HA found in connective tissues [169]. A summary of the concentration distributions and the average MW of HA in various healthy human tissues is in Table 3.
Table 3.
HA concentration and molecular weight in human tissues and fluids.
| Tissue | Concentrationa | Molecular Weight |
|---|---|---|
| Skin (total) | 440–520 μg/g wet weight, mostly in dermis [177] | 2000–5000 kDa [178] |
| Synovial Fluid | 40–3800 μg/mL [179,180] | Majority 6000–7000 kDa [181] |
| Vitreous Body of the Eye | 8–400 μg/mL [182,183] | 2000–4000 kDa [184] |
| Umbilical Cord (Wharton’s jelly) |
20,000 μg/g [185] | 1100 kDa, 700 kDa during acute funisitis (inflammation of umbilical cord), and 520 kDa in necrotizing funisitis [186] |
| Amniotic fluid | 20 μg/mL (16–20th week of pregnancy), 1 μg/mL in the 30th week until the end of pregnancy [187] | 330 kDa (16th week of pregnancy), mixture of >1000kDa and <100kDa (40th week) [188] |
| Aqueous Humor | 1.1 μg/mL [189] | 1000–5300 kDa [190] |
| Lymph Fluid | 0.1–18 μg/mL [191] | 1400 kDa, large range [191] |
| Human Milk | 0.8 μg/mL (immediately postpartum), 0.2 μg/mL (60 days after birth) [192] | 440 kDa [193] |
| Blood plasma | 0.06–0.7 μg/mL[191] | 140–270 kDa [191,194] |
Tissue concentrations are reported in units of μg HA/g of wet weight of the tissue, and fluid concentrations are reported in units of μg HA/mL of the fluid of interest.
2.1.2.4. Synthesis and Degradation
Hyaluronan synthases (HAS) are the enzymes responsible for the production of HA. There are three known isozymes, HAS1, HAS2, and HAS3 [154]. The isoenzymes lengthen the HA polymer by repeated addition of glucuronic acid and GlcNAc groups [154]. Only one of these enzymes is necessary for the production of HA, but their varying kinetics and concentrations result in the production of various MWs [195]. HAS1 and HAS2 are moderately active and produce high MW HA, whereas HAS3 is the most active and polymerizes low MW HA [154]. The presence of certain growth factors, including epidermal growth factors, and cell types, such as keratinocytes, increase HA synthesis. HAS enzymatic activity can be impeded by both mannose [196] and natural antisense mRNA complementary to HAS2 [197].
HA is degraded into monosaccharide components by enzymes, such as hyaluronidases, chondroitinases, and hexosaminidases, as well as by reactive oxygen species (ROS) [198,199]. Two main hyaluronidases, Hyal-1 and Hyal-2, cleave HA in somatic tissues [200]. Hyal-2 degrades high MW HA into intermediate size fragments of ~20 kDa [201]. Inside the cell, the HA fragments are further degraded by Hyal-1 into oligosaccharides (predominantly tetrasaccharides) [202]. The degraded HA fragments and oligosaccharides participate in various signaling cascades described in Section 2.2.2 [202]. There are two classes of enzymes, endo- and exoglycosidases, that cleave larger fragments in the center of the molecule and the termini, respectively [200]. Specifically, hyaluronidases are endoglycosidases that hydrolyze the β−1,4 or the β−1,3 linkage in the center of HA molecules depending on the type of hyaluronidase [200,203]. The oligosaccharide products are further degraded by the exoglycosidases β-D-glucuronidase and β-N-acetyl-glucosaminidase, which remove terminal sugars [200,204,205]. Stern et al. provide a thorough review of HA degradation methods [200].
2.2. Interactions between collagen/HA and cells
Both collagen and HA play major roles in the development and maintenance of the ECM and also provide biochemical and physical cues to cells for functions such as adhesion, proliferation, differentiation, and production or degradation of the ECM. The interaction of collagen and HA with cell surface receptors contributes to matrix remodeling, including the synthesis of new molecules, such as collagen and proteoglycans, and the production of degrading enzymes. Additionally, the degradation of collagen and HA into smaller fragments can trigger other cell responses. Collagen fragmentation can lead to the recruitment of neutrophils, monocytes, macrophages, mast cells, and fibroblasts. Collagen and HA also contribute to the progression and the mitigation of various diseases such as cancer and osteoarthritis [206–208].
2.2.1. Collagen & cells
Collagen interacts with cells largely via various integrin cell surface receptors. Collagen binds to integrins that contain the β1 subunit and one of the following alpha subunits: ɑ1, ɑ2, ɑ10, and ɑ11 [209]. In native collagen, the minimal binding sequence for integrins includes GFOGER and GROGER [6,210,211]. Additional integrins can bind to hidden RGD sites that become available when collagen is denatured [211]. Denatured collagen fragments can also temporarily anchor to the ECM and provide additional sites for cellular migration and adhesion [212]. KGD motifs can also be recognized by RGD-binding integrins but with different affinity than for RGD [210]. Fibrillar collagens have a strong avidity for the ɑ2β1 integrin, which regulates cell adhesion and migration. Collagen binding with integrins is reviewed extensively by Heino et al. [210].
Other surface receptors that collagen interacts with include dimeric discoidin receptors (DDRs), DDR1 and DDR2, glycoprotein VI (GPVI) on platelets, and leukocyte-associated immunoglobulin-like receptor-1 (LAIR-1) on immune cell surfaces [6,207,212]. DDRs can bind to the GVMGFO motif in collagen. DDRs regulate cell functions such as proliferation, differentiation, and matrix maintenance [212]. GPVI and LAIR-1 bind to GPO motifs, which are found on all collagens [210]. GPVI is an important receptor found on platelets, and collagen binding can trigger signaling events and activate the function of platelets during thrombosis. Interactions between LAIR-1 and collagen can downregulate immune responses as LAIR-1 is a surface receptor found on peripheral blood mononuclear cells. Thus, collagen is implicated in numerous autoimmune diseases [207].
NC domains found in collagen offer an additional site for binding to cell surface receptors and thus play a pivotal role in numerous cellular processes. For example, NC1 in various collagens (e.g., Col IV, Col XVIII) interacts with integrins on endothelial cells (ECs) and inhibits angiogenesis [213]. The importance of NC domains in the molecular architecture of the ECM and its biological functions are highlighted by Ortega and coworkers [214].
2.2.2. HA & cells
HA interacts with cells in numerous pathways through surface receptors to initiate cell functions. CD44 is the primary class of cell surface receptors that interact with HA [206]. CD44 is a transmembrane glycoprotein found throughout the body on numerous cell types, such as leukocytes, fibroblasts, mesodermal cells, and cancer cells [215]. HA interactions with CD44 vary based on cell type. These interactions have been shown to play a role in cell functions such as cell adhesion, metastasis, endocytosis, cell signaling, cytokine release, and matrix deposition [206,216]. Signaling pathways triggered by CD44 binding include tyrosine kinases (p185 and c-Src), Rholike GTPases, and Rac1 signaling [217]. CD44 recognizes HA via various interactions such as hydrophobic interactions and numerous hydrogen bonding sites [218].
Receptor for HA-mediated mobility (RHAMM) is another major receptor for HA binding. HA interacts with the B(X7)B motif found in RHAMM, where “B” is a basic amino acid, excluding histidine, and “X” includes at least one basic amino acid and excludes acidic residues [219]. HA RHAMMs are found on the surface of cells, in the cytosol, and in the nucleus and activate a number of signaling cascades [217]. Finally, HA interacts with the hyaluronan receptor for endocytosis (HARE), which aids in clearing 80–90% of total HA [199,206].
The interaction between cells and HA has also been shown to depend on the molecular weight of HA [198,217]. In particular, studies have shown that low MW HA fragments can lead to the expression of inflammatory genes in a variety of cell types [198,203]. High MW HA fragments have been shown to activate protein-tyrosine kinase pathways in ECs and Ras-transformed fibroblasts [217]. One potential way for cells to discern various chain lengths of HA is that CD44 receptors can cluster and crosslink together to bind to numerous binding sites along a single HA molecule [203,216]. When smaller HA fragments (oligosaccharides of ~6–18 sugars) competitively displace larger HA-CD44 complexes, it signals an unstable complex and the cellular response may come to a halt [203,220]. An alternative rationale for the differences in the effect of different molecular weights is that size may impact the uptake of HA, and therefore intracellular signaling is an important factor [203].
HA is degraded into different MW fragments by the mechanisms detailed in Section 2.1.2.4. The degradation of HA is a necessary step to signal that homeostasis has been perturbed [198,217]. When HA is cleaved into smaller fragments (HA <1 MDa or HA oligosaccharides), these fragments can lead to scar tissue formation or induce an immune response [198,199,221]. HA is largely cleared from circulation via endocytosis in the lymph nodes and liver by HARE receptors [199]. Finally, another method for altering the cell signaling dynamics is for chondroitin sulfate (CS) to be replaced by HA via trans-esterification or non-covalent bonds to heavy chains [222]. When the exchange of CS for HA occurs, HA can covalently bind to the surface of cells and participate in the formation of additional pericellular matrix [222,223].
2.3. Summary and future outlook of collagen and HA function
Collagen and HA are biomolecules found in the ECM and provide mechanical stability and biochemical and biomechanical cues to cells. Collagen is the most abundant protein in the human body and forms crosslinked fibrous networks in the ECM. HA is a negatively-charged, non-sulfated GAG and provides crucial signals for biological processes, including wound repair and inflammation. Both collagen and HA interact with surface receptors of cells to induce cell activities such as migration, proliferation, and matrix remodeling. Thus, collagen and HA confer desirable properties for biomedical applications including regeneration of cartilage, bone, and skin. Currently, most research focuses on collagen I and HA scaffolds, which offer platforms to decouple and study complex biological processes. In the future, creating blends consisting of numerous collagen types may create more efficacious medical treatments as more than one type of collagen is often present within native tissue. Furthermore, these scaffolds can help uncover the biological effects of HA molecular weight and CD44 interactions with HA since these factors are not yet fully characterized for various tissues.
3. Sources and commercial applications of collagen and HA polymers
Collagen can be naturally sourced or synthetically developed. Shorter sequences of collagen, including collagen mimetic peptides [224–227], collagen-like proteins found in bacteria [228–232], and hydrolyzed collagen peptides [233] have also been developed into biomaterials; however, this section will focus on full-length collagen from animal sources and recombinantly-produced collagen developed in various host organisms due to their ubiquitous use in the biomedical field. HA is commercially sourced from animals and microorganisms. This section will focus on these commercial sources as well as the use of recombinant microorganisms and cell-free methods to produce HA. Finally, the commercial applications of collagen and HA polymers will be discussed.
3.1. Sources and characteristics of different types of collagen
3.1.1. Animal-derived collagen
As the most abundant mammalian protein [6], collagens, specifically collagen I, can be sourced from the majority of animals. Mammalian collagens are of interest due to the high level of conservation in the triple helical region across species [234–236] and the presence of post-translational modification machinery that confers the biochemical functions of collagen [237,238]. The major sources of collagen for scientific research are highly collagenous tissues, such as skin, tendons, bones, and cartilage, derived from cows, pigs, and sheep [239]. The food industry provides a bountiful supply of tissues for collagen extraction without extra cost, and value is added to previously polluting byproducts. This practice has resulted in high yields of low-cost collagen that is similar to human collagens and ideal for biomedical research. Collagens from equine, murine, and avian sources are also common and have been characterized and studied for biomedical applications [239]. More exotic sources, which include kangaroo tail tendon [240], alligator bone [241], and frog skin [242], highlight the abundance of the protein.
Collagen extraction protocols consist of four general steps: 1) raw material separation and size reduction, 2) removal of non-collagenous components, 3) acid or enzymatic collagen extraction, and 4) purification by salt precipitation or chromatography methods [29,243,244]. Crosslinked collagen derived from animals is not water soluble; thus its extraction protocols involve chemical and enzymatic reactions to increase the solubility of the protein. Organic acids, most commonly acetic acid, are used to break the non-covalent inter- and intramolecular bonds and increase the solubility of the protein [29] to yield acid-soluble collagen. Acid solubilization can be followed by enzymatic extraction, which improves the yield of the protein [245]. Pepsin-soluble collagen is formed by using a non-specific collagen enzyme, typically pepsin, to increase solubility by cleaving the telopeptide regions on the ends of the triple helical structures, which are major sites of interchain cross-links [29]. The method of extraction results in distinct polymerization profiles (Fig. 3), subtle architectural differences in fibril network organization and fibril–fibril interactions, and differences in mechanical integrity [243,245].
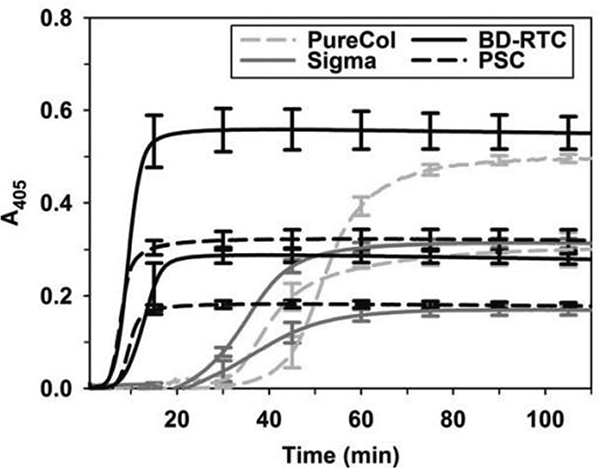
Fig. 3.
Collagen origin and processing affects polymerization kinetics. Polymerization kinetics of “in house” extracted porcine skin collagen (PSC) extracted using acid compared to commercially sourced acid solubilized bovine dermis (Sigma), acid solubilized rat tail (BD-RTC) and pepsin treated bovine dermis (PureCol) collagens. Collagen was polymerized at concentrations of 0.5 and 1.0 mg.ml (lower and upper curve for each collagen source, respectively) and measured spectrophotometrically at 405 nm. Reproduced with permission from Kreger et al., 2010 [245]. Copyright 2010 Wiley Periodicals, Inc.
Despite animal-derived collagen leading to an abundance of low-cost material, variabilities due to animal species and choice of tissue contribute to variations in material properties. Recently, there has been increased interest in characterizing collagen material properties from different animal species [112,246–249]. Differences in material properties, including ultimate stress, ultimate strain, and toughness, have been observed [246]. Microstructural architecture also appears to vary with origin species [247,248], and there are pore size variations as large as 51.6% in chitosan-collagen crosslinked gels based on species (Fig. 4) [247]. Collagen from tissues with strict hierarchical organizations, such as equine tendons, results in materials that retain partial lateral packing and have higher mechanical properties [112] compared to other sources. In addition, compared to collagen from young mice, collagen originating from older mice formed fibrils more slowly, generated fibrils with smaller diameters, and resulted in networks that were less dense [250]. Despite these material variabilities, collagen from young or old mice resulted in scaffolds that promoted cell adhesion; however, ECM deposition amount, and gel contraction varied [250]. Comparison studies of collagen from different species often fail to consider the age of the animal, extraction methods, and tissue of choice, and thus conclusive generalizations are difficult to make.
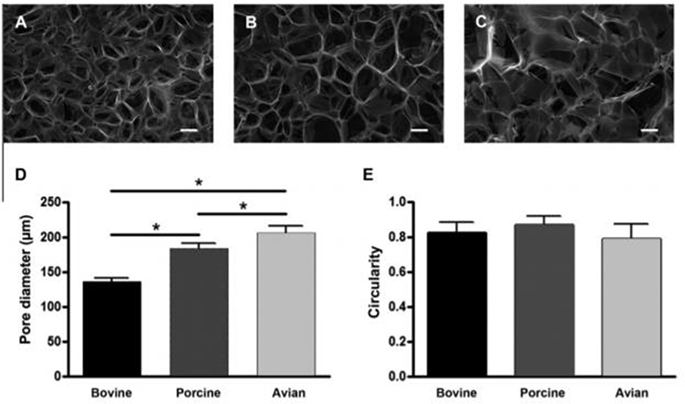
Fig. 4.
Animal source of collagen impacts the microstructure of polymerized scaffolds. Scanning electron microscopy images of collagen–chitosan scaffolds made with (A) bovine, (B) porcine, or (C) avian collagen (scale bar represents 100 μm). (D) Average pore size of scaffolds from bovine, porcine or avian sources. Give the same fabrication process, collagen source results in significant differences in pore size. (E) No significant differences were observed in the circularity of pores. Reprinted from Acta Biomaterialia, 7, R. Parenteau-Bareil, R. Gauvin, S. Cliche, C. Gariépy, L. Germain, F. Berthod, Comparative study of bovine, porcine and avian collagens for the production of a tissue engineered dermis, 3757–3765, Copyright 2011, with permission from Elsevier [247].
Concerns about safety of materials sourced from animals prevent full clinical translation. The incidence of adverse reactions to acellular collagen implants are rare, but they do occur [251]. Animal sources, especially bovine, carry inherent risk of disease transmission, including bovine spongiform encephalopathy. Bovine collagen is used widely, but diseases originating from bovine collagen can be dormant for decades before the appearance of the first symptoms [247]. Clinical observations indicate that between 2–4% of the population possess an allergy to bovine collagen I [252–255]. An additional 1% develop an allergy to bovine collagen postoperatively [251]. Granuloma and localized inflammation have been observed in rare cases and resolved within a year [251].
Although the triple helical regions are mainly conserved across species, the amino acid sequences of the terminal regions exhibit ≤50% variation between species [256]. Speculation that this region is a source of adverse immune reaction led to the fabrication of atelocollagen, which is collagen treated with pepsin to remove the N- and C- terminal telopeptides. The lack of telopeptides, however, alters nucleation and fiber structure [257,258]. Collagen I is not typically associated with an autoimmune response. However, studies implicated collagen II as a potential auto-immunogen [251,259–261]. For a detailed review of collagen immunogenicity, see the 2004 review by Lynn et al. [251].
Religious and cultural beliefs further prevent clinical translation of bovine and porcine collagens. In addition, pepsin, an enzyme crucial for the extraction of collagen from animal sources and the production of atelocollagen, is typically of porcine origin and raises further cultural concerns [29]. The safety and religious concerns about bovine and porcine collagens have paved the way for marine-sourced collagen as an alternative.
Marine animals are typically associated with less religious and cultural significance and can be readily sourced from the food industry. Marine sponges and jellyfish [29,262] are the most widely studied marine invertebrates for collagen source as they are cultivated for their pharmacologically active terpenoid and alkaloid compounds [29,263]. Other invertebrates that have been studied include cuttlefish, sea anemone, prawn, starfish, jellyfish, sponge, sea urchin, octopus, squid, and mollusk [29]. Collagen derived from aquatic species only slightly differs in amino acid composition from mammalian sources [29]. However, some sources lack imino acids compared to human collagen. These molecules are crucial for the structural integrity of collagen fibers; thus, their absence leads to denaturation temperatures as low as 16–20 °C [29]. The low denaturation temperature is a concern as clinical translation will require the material to remain intact at human physiological conditions. Collagen extracted from carps, tilapia, and few other warm water fish have denaturation temperatures between 32–37 °C and thus are better suited for biomedical applications [29]. Finally, marine collagens are preferable to bovine collagens from a cultural perspective. For a detailed review of marine collagens, see these review papers [29,33].
Despite source variability, cultural and religious concerns, and safety issues, animal-derived collagen remains the gold standard material for tissue engineering applications. Animal-derived collagen is abundant in nature, can be extracted from food and pharmaceutical byproducts, and resembles human collagen in biomechanical and biochemical aspects. Thus, animal-derived collagen is widely available and accessible for research and clinical purposes.
3.1.2. Recombinant human collagen
Given the drawbacks of animal-derived sources of collagen, recombinant human collagen offers an alternative means for developing biocompatible collagen with low variability and immunogenicity. Proteins are recombinantly produced by cloning genetic material into a host that produces the protein. Recombinant collagen is particularly attractive for the production of collagen types other than collagen I because sourcing and purification of less common collagen types are difficult.
Researchers have developed recombinant human collagen, but its production needs to achieve: 1) scalable and inexpensive production that can compete with animal-derived sources, 2) human collagen levels of post-translational modifications and higher order structural arrangement, and 3) heterotrimeric collagen expression in addition to homotrimeric expression [264]. Different host organisms have been developed to address these challenges (Table 4).
Table 4.
Advantages and disadvantages of different hosts in recombinant human collagen expression and production.
| Host | Advantages | Disadvantages | Collagen Types |
|---|---|---|---|
|
Bacteria • Escherichia coli [265,266] |
• Inexpensive • Scalable • Well-characterized |
• Poor transformation efficiency of long plasmids • No native enzyme for post-translational modification |
III [265,266] |
|
Yeast • Saccharomyces cerevisiae [267–270] • Pichia pastoris [271–274] |
• Low maintenance cost • Well-characterized |
• No native enzyme for post-translational modification | I [267,270,271] II [272] III [268,269,273,274] |
|
Plants • Tobacco [275–277] • Maize [278] |
• Good transformation efficiency of long plasmids • Native prolyl hydroxylase activity • Scalable |
• Low native enzyme hydroxylation activity | I [275–278] |
|
Mammalian cells • Chinese hamster ovary cells [279] • HeLa cells [280] • human embryonic kidney 293 cells [281–283] • Fibrosarcoma HT 1080 cells [283–285] |
• Simple cloning schemes • High cloning efficiency • Native hydroxylase and lysine glycosylase activity |
• Poor yield • Costly to culture • Long production times |
I [284] II [285] IV [279] V [281] VII [282] X [283] XII [280] |
|
Insects • Spodoptera frugiperda Sf9 cells [286–290] • Drosophila melanogaster S2 fly cells [291] • Bombyx mori silkworms [290,292,293] and silkworm cells [290] |
• Established transfection protocols • High density culture for scale-up • Cheaper scale-up than mammalian cells • Native hydroxylase activity |
• Low native enzyme hydroxylation activity | I [286,292] II [290] III [287,288,293] IX [289] XXI [291] |
Yeast and bacterial recombinant hosts are well characterized and inexpensive; however enzymes required for post-translational hydroxylation and glycosylation must be transformed into these hosts [266,294]. More complex systems, such as plant, mammal, and insect cell hosts, have native enzymes for post-translational modification, but yield and enzyme activity are low. Often, recombinant collagen constructs result in truncated polypeptide chains due to native host protease degradation or codon usage bias [295]. Overall, yield and native post-translational modifications continue to elude the recombinant collagen community to achieve true mimetics of human collagen, but several groups have used recombinant human collagen to develop novel biomaterials [296–300].
3.2. HA sources and property variation
3.2.1. Commercial extraction methods of HA production
The bioactivity of HA and its possible applications are highly dependent on the polymer molecular weight [155,301]. It is difficult to obtain high MW HA due to degradation of HA during extraction, purification, storage, and sterilization [302,303]. HA is sensitive to harsh processing conditions such as extreme pH, high temperatures, and long durations of heating [304]. Extreme acidic conditions disrupt hydrogen bonding and can lead to random polymer degradation and a reduction of viscosity. If thiols or transition metals are present, they can cause the production of reactive hydroxyl radicals that cleave HA [160]. If HA is produced with residual hyaluronidase, it is susceptible to enzymatic degradation [305]. These chemical, thermal, and enzymatic factors can result in lower MW HA products with wide polydispersity.
HA, unlike collagen, is structurally preserved across species [306]. Therefore, HA extracted from other species is chemically identical to human HA. It was initially extracted from sources, such as rooster combs (7.5 mg HA per g of tissue) and human umbilical cords (4.1 mg/g), that have high HA concentrations [302,307,308]. High MW HA (~1 MDa) can be derived from tissues [155] and is useful in ophthalmological, orthopedic, and tissue engineering applications [301,309,310]. However, the yield of animal-sourced HA is limited by the naturally occurring concentrations in tissues, and adverse immune reactions can occur from any proteins, proteoglycans, or DNA remaining from the extraction and purification processes. These limitations have decreased the use of animal-sourced HA.
3.2.2. Production of HA based on microorganisms
HA was commercially produced from several microorganisms after HA synthase (HAS) operons were identified in Streptococcus bacteria in the 1990s [311,312]. HAS enzymes from Streptococcus species extend the HA chain from the reducing end and are membrane-bound, which is similar to human HAS enzymes [307]. The ideal host organism should be able to efficiently synthesize high MW HA, be non-pathogenic, and have no hyaluronidase activity [302].
Initial commercial production was performed with Streptococcus equi and Streptococcus zooepidemicus, and it was found that S. equi natively produce HA with a lower MW than S. zooepidemicus [160]. Groups have been investigating more efficient strains and production/purification systems for these Streptococcus strains [313–320]. For example, wild-type production of HA from S. zooepidemicus produced HA MWs of ≤1.8 MDa, whereas engineered strains produced MWs ≤3.4 MDa [321].
Numerous non-pathogenic microorganisms, including Bacillus subtilis [322,323], Lactococcus lactis [324,325], E. coli [326–328], and others [329–333], have been genetically manipulated to produce HA at a research scale [307,334]. B. subtilis is a commonly used industrial producer and is generally recognized as safe (GRAS). It has native enzymes homologous to several HAS enzymes in Streptococcus, and other necessary HAS genes can be introduced through transformation [322,323,325,335,336]. L. lactis has also been used for its GRAS status and lack of hyaluronidases [324,337,338]. Many microorganisms, such as E. coli, lack native HAS genes, so genes from HA-producing bacteria must be transformed into the hosts [326–328].
There are several limitations inhibiting the progress of microorganism production of HA. Large scale fermentation of HA is difficult due to increased viscosity as HA concentration increases. High viscosity prevents proper mixing and results in a poor oxygen transfer rate, which reduces yield [339]. Researchers characterized in vitro and in vivo HA synthesis from different bacterial strains and determined that the maximal HA MW is specific to an individual synthase, but shorter HA chains may result due to physiological and metabolic factors and production conditions [321,340,341]. Polydispersity in the MW of the final product is still an issue as the distribution is heavily controlled by reaction stoichiometry and other culture conditions. Finally, there is the possibility of endotoxins from pathogenic microorganisms or bacterial contamination from binding proteins and nucleic acids that may elicit a human immune response [334]. Compared to HA extraction from animals, bacterial production commonly results in shorter HA chain length but higher yield.
3.2.3. Cell-free methods of HA production
To avoid several of these drawbacks of microorganism fermentation, HA has also been produced at the research-scale in cell-free in vitro systems that utilize purified HAS enzymes. Although most species produce HAS enzymes that need to be tightly bound to the cell membrane, Pasteurella multocida produces HAS enzymes that do not require membrane association and can synthesize HA in a cell-free system [342–344]. Researchers are also making progress to develop the more ubiquitous membrane-bound HAS enzymes for cell-free production [345].
Though non-pathogenic hosts and cell-free systems offer alternatives to current commercial production, these systems do not have the capacity to produce HA at a commercial scale. All current commercial and experimental methods are limited in their ability to efficiently produce high MW HA with a narrow molecular weight distribution at a large scale. The increase in use of HA in the biomedical industry will continue to drive future innovation to optimize HA production.
3.3. Commercial applications of collagen and HA polymers
Due to their biological activity and relative abundance, both collagen and HA polymers have been used in a variety of biomedical commercial applications. To meet the requirements of their intended biomedical uses, the polymers are often modified and processed to create various forms. Additional factors, including other biopolymers and growth factors, are added to work in conjunction with collagen or HA for the intended biomedical use. While both collagen and HA have been used extensively, few products incorporate both in combination. As such, this section will detail several commercial applications of collagen and HA, including orthopedic, ophthalmic, and surgical applications.
3.3.1. Collagen matrices and sponges
Collagen matrices are widely used for clinical applications ranging from skin grafts and wound dressings, to regenerative orthopedic applications. Though the specifics may differ between manufacturers, the general procedure for creating collagen matrices involves dissolving previously-extracted collagen at high concentrations, then freezing and lyophilizing the material before commercial distribution. Further processing of this matrix through extensive chemical crosslinking results in a collagen sponge, which is able to absorb large quantities of water [346]. Most commercial collagen products are primarily composed of collagen I due to its greater availability compared to other collagen types.
3.3.1.1. Collagen matrices for wound dressings and skin healing
Collagen matrices have been produced by several companies as platforms for treating burns and chronic skin wounds. By retaining water and facilitating cell migration and proliferation, collagen matrices maintain a moist environment at the wound site and promote wound healing. Manufacturers often add other components to collagen sponges to augment bioactivity and effectiveness. Examples include Integra Dermal Regeneration Template Single Layer (Integra LifeSciences, USA), which is a collagen sponge with GAGs [347,348], and Matriderm (MedSkin Solutions Dr. Suwelack AG, Billerbeck, Germany), which is a collagen matrix with elastin [347,349,350]. Although these products are sold as acellular scaffolds, some manufacturers sell collagen-based skin graft products, such as Apligraf (Organogenesis, USA), that are pre-cultured with dermal fibroblasts several weeks prior to cryopreservation and packaging [351–353].
3.3.1.2. Collagen matrices for orthopedic applications
To treat critical-sized bone defects, collagen sponges have been used to provide a scaffold for the growth of new bone. The INFUSE bone graft (Medtronic, USA) is a collagen sponge loaded with recombinant human bone morphogenetic protein 2 (rhBMP-2) [354]. This collagen sponge delivery vehicle has been used in several orthopedic applications including the filling of craniofacial bone defects and spinal fusions to treat degenerative discs [355]. Collagen-based fillers or scaffolds such as Collagraft (Zimmer Biomet, USA), Collapat (Zimmer Biomet, USA), Healos (DePuy Synthes, USA), Infuse (Medtronic, USA), etc. have US Food and Drug Administration (FDA) approval for use in orthopedic treatments [356,357].
For the treatment of critical-sized cartilage defects, collagen matrices have been used as a delivery vehicle for matrix-assisted autologous chondrocyte implantation (MACI) procedures. Despite cartilage consisting primarily of collagen II, most collagen matrices for MACI procedures are made of collagen I. Some matrices, such as Chondro-gide (Geistlich Pharma, Germany), Maix (Matricel, Germany), and MACI (Verigen, Germany), are a blend of collagen I and III [358,359]. These collagen matrices are often made into thin sheets and secured into place using an adhesive such as fibrin glue.
3.3.2. Collagen sutures
Resorbable sutures are absorbed over time and eliminate the need for removal by a clinician after wound healing. Catgut sutures are made by twisting strands of animal-derived collagen together [360], and a patient’s own proteolytic enzymes are capable of degrading the suture over time. This process is much faster than the degradation of synthetic polymers, such as poly(lactic-co-glycolic acid), that hydrolyze slowly and hold tensile strength for much longer [360,361]
3.3.3. Recombinant collagen biomaterials
Commercial recombinant human collagens are regularly used in biomedical research. Fibrogen commercially developed recombinant human collagen from yeast [232]. A group of researchers used this collagen to develop a matrix for the treatment of myocardial infarction [362]. Another group used Fibrogen recombinant collagen II for the chondrogenesis of mesenchymal stem cells (MSCs) [363]. Fujifilm developed a commercial recombinant collagen peptide based on collagen I. This material has been used to improve culture conditions for stem cells [364–367], to mitigate slowing of cardiac conduction by stromal cells [368], and to serve as a bone graft material [369].
3.3.4. HA solutions
One common treatment for osteoarthritis is the supplementation of the patient’s synovial fluid with the injection of high MW HA. To further increase the viscosity of the hyaluronic acid, the polymer may be mildly crosslinked, such as in Synvisc (Sanofi, France) [370]. However, many commercial products are sold as unmodified hyaluronic acid or sodium hyaluronate. Despite its wide-spread use, the efficacy of synovial fluid viscosupplementation is still disputed [371].
Because HA is a natural component of the vitreous humor, solutions of HA have also been manufactured for ophthalmic applications. HA solutions have been used to aid in the implantation process of intraocular lenses following cataract surgeries [372]. However, because of its high hydrophilicity, HA has been found to adversely increase intraocular pressure following procedures, and work is being done to mitigate this side effect [373].
For soft tissue reconstruction or plastic surgery purposes, HA solutions can be injected subcutaneously as a dermal filler. To prolong its lifetime after injection, the polymer chains can be modified to prevent enzymatic degradation by hyaluronidase. An example of a modified product is HYADD3 (Fidia Advanced Biopolymers, Italy), where a dodecylamine is conjugated to the carboxyl group for steric hindrance, which slows down enzymatic degradation [374].
3.3.5. HA for wound healing
To create HA-based skin grafts and wound dressings, many manufacturers have modified the polymer, such as through benzyl esterification, to reduce its solubility in water [375]. The degree of modification can be modulated to control the degradation properties of the material. Commercially, benzyl-esterified HA is sold under the name HYAFF, with different grades depending on the degree of modification [375]. This material can then be processed into bulk products. For example, HYALOFILL (Anika Therapeutics, USA) is a soft fabric-like material made from uncrosslinked HYAFF fibers and is used for wound dressings [376]. Composite material wound dressings have also been developed using HYAFF, such as the Hyalomatrix wound dressing (Fidia Advanced Biopolymers, Italy), which combines uncrosslinked HYAFF fibers with a silicone membrane for added stiffness [377].
3.3.6. Regulation of commercial products
Currently, the FDA treats many of these collagen- and HA-based products as medical devices. General FDA guidance conforms to the ISO standard 10933–1:2018: Biological Evaluation of Medical Devices, which outlines the various biocompatibility tests for evaluation of medical devices [378]. Tests include an examination of cytotoxicity, material-mediated pyrogenicity, acute and chronic system toxicity, carcinogenicity, and the effects of implantation. Other standards, such as ASTM F2212–20: Standard Guide for Characterization of Type I Collagen as Starting Material for Surgical Implants and Substrates for Tissue Engineered Medical Products (TEMPs) (ASTM International, F2212–20) [379] and ASTM 2150: Guide for Characterization and Testing of Biomaterial Scaffolds Used in Tissue-Engineered Medical Products (ASTM International, ASTM F2150 – 19)[380], provide guidance on how to test the biocompatibility of these materials. In general, collagen- and HA-based materials meet these guidelines due to their natural bioactivity and degradability [381]. However, care must be taken with regard to the processing of the materials to ensure they do not contain toxic contaminants, are sterile after processing, and that modifications do not elicit adverse effects upon application. For new products, premarket approval is required by the FDA to ensure that the product is safe to use and operates as intended, based on both laboratory and clinical data. However, in cases where it can be proven that the product is similar to one already on the market, the FDA approval process can be expedited through the premarket notification path, also known as a 510(k). This approval pathway allows for products similar to those whose safety and efficacy have already been proven to reach the market significantly faster.
4. Fabrication of collagen and HA hydrogels
Crosslinking, chemical modifications, and environmental conditions such as pH and temperature can be used to tune the properties and functionalities of collagen and HA hydrogels. These hydrogels are usually designed by modulating various physical and chemical parameters to achieve the desired properties suitable for specific applications, which are discussed in detail in Section 6. Discussed below are some of the specific factors and modifications that can be used to tune the properties of collagen, HA and ColHA hydrogels.
4.1. Fabrication parameters and polymerization conditions for collagen hydrogels
Collagen fibrillogenesis and polymerization into a hydrogel depend on different parameters such as the collagen source, concentration, pH, temperature, and ionic strength. These factors regulate the hydrogen bonding, hydrophobic interactions, and electrostatic interactions between amino acid residues and control polymerization rate as described below [382,383].
4.1.1. Collagen source and solubilization method
Animal source (e.g., species, age), tissue source (e.g., skin, tendon), and extraction methods (e.g., acid-solubilized, pepsin-digested) play important roles in collagen polymerization kinetics and hydrogel properties. Kreger et al. reported that, under identical polymerization conditions, acid-solubilized porcine skin collagens polymerize more quickly due to intact telopeptide regions as compared to pepsin-digested bovine or porcine collagens [245]. Wolf et al. found that gels of pepsin-digested collagens have longer fibrils and larger pores compared to acid-solubilized collagen gels, which have high fibril density, shorter fibrils, and smaller pores [243]. The age of the animal used for collagen extraction also plays a vital role in polymerization. Collagen from older animals resulted in more malleable hydrogels with smaller-diameter fibrils and a lower density of fibrils [250].
4.1.2. Collagen concentration
Increasing the collagen concentration in hydrogels reduces the diffusion coefficient of molecules diffusing within the hydrogels [384]. Ramanujan et al. demonstrated this inverse relationship by using fluorescence recovery after photobleaching to measure diffusion coefficients of dextrans with various molecular weights (4–2000 kDa) in collagen hydrogels. Similar results were reported with decorin- or HA-supplemented collagen hydrogels containing 2000 kDa dextran [385] and collagen-alginate hydrogels with 3–500 kDa dextran [386].
Collagen concentration also affects hydrogel properties. For example, an increase in concentration resulted in hydrogels with higher fibril density and reduced pore size [387,388]; however, fibril diameters were unaffected by changing collagen concentration only [245]. Several investigators have also reported a positive correlation between collagen concentration and the shear and elastic moduli of the hydrogel [245,389,390]. Fraley et al. however, documented that, the elastic modulus decreased when collagen concentration increased from 1 to 1.5 mg/mL and then increased when the concentration increased from 1.5 to 2.5 mg/mL [391]. Increasing the collagen concentration from 1 to 1.5 mg/mL also decreased the alignment of collagen fibers without affecting the average pore size, whereas increasing from 1.5 to 2 mg/mL drastically reduced the pore size and slightly decreased the fiber alignment.
4.1.3. pH
pH has a significant influence on the resulting physical and mechanical properties of collagen hydrogels and depends on various factors such as the ratio of collagen to neutralization agent, collagen dilution ratio, type of buffer, and collagen concentration [392]. An increase in pH results in collagen networks with longer and thinner fibers and increased pore density with reduced pore area fraction and size [393,394]. Furthermore, the rigidity of collagen gels increases with an increase in pH [395].
4.1.4. Temperature
At higher temperatures, the self-assembly of collagen molecules occurs more rapidly due to a larger number of hydrophobic and electrostatic interactions that increase fibril precipitation while limiting lateral aggregation and result in fibers with a reduced number of bundled fibrils [396,397]. Higher polymerization temperature, therefore, results in a less-ordered structure with altered mechanical and transport properties. Yang et al. varied the polymerization temperature from 4 °C to 37 °C and observed that lower polymerization temperatures resulted in enhanced bundling of fibrils that resulted in increased fiber diameters, larger pore sizes, and, consequently, better proliferation and migration of glioma cells [396,397]. Chrobak et al. emphasized the role of temperature on gel stability by creating more stable and less degradable microchannels in gels polymerized at room temperature compared to gels formed at 37 °C [398].
4.1.5. Ionic strength
The structural and mechanical properties of collagen hydrogels are highly influenced by the ionic strength of the polymerization solution. Gobeaux et al. demonstrated this effect by polymerizing gels at ionic strengths ranging from 24 to 1300 mM and reported variations in hydrogel structural and optical properties [387]. Even with a smaller range (64.2 to 174 mM), Achilli and Mantovani found that variations in ionic strength impacted the mechanical properties of gels [399]. They also showed that the dependence of the compression modulus on ionic strength is interdependent on pH and temperature. Wood and Keech noted that collagen fibril diameter generally increased at higher ionic strengths, and that, at lower ionic strengths, collagen self-assembly was faster and resulted in a smaller number of bundled fibers [400].
4.1.6. Macromolecular crowding agents
Macromolecular crowding is a phenomenon in which high concentrations of macromolecules occupy space and generate excluded volume effects [401]. The assembly of collagen in solution is driven by diffusion-limited growth of nucleated monomers [402]. Crowding during collagen polymerization can impact the fibril architecture of collagen in hydrogels. Many macromolecules, such as Ficoll, HA, polyvinylpyrrolidone, and poly(ethylene glycol) (PEG), have been studied as crowding agents in collagen hydrogels [403–406]. For example, PEG is a commonly used biologically inert macromolecular crowding agent that, when added, results in tighter and less degradable collagen fiber networks [407]. Adjustments to the amounts of PEG added during collagen assembly modulated fibril length and pore size without changing collagen density and hydrogel stiffness.
4.1.7. Summary of fabrication and polymerization parameters of collagen hydrogels
All the polymerization and fabrication parameters summarized above have a significant impact on single or multiple facets of collagen fiber structure. These factors can thus be modulated to form hydrogels according to specific requirements. For example, hydrogels with reduced fiber diameter and small pore size can be achieved by increasing pH or temperature or by decreasing ionic strength. Antoine et al. provides an excellent review on the quantitative characterization of hydrogel properties and their correlation with fabrication parameters [392].
4.2. Chemical crosslinking of collagen hydrogels
The functional groups of polymer chains can be crosslinked, or connected to one another through physical or covalent bonding, to provide structural stability. An ideal crosslinker not only provides stability with improved mechanical performance but also should be non-cytotoxic for biomedical applications. Crosslinking of polymer chains affects physicochemical and biochemical properties of scaffolds such as susceptibility to chemical and enzymatic degradation, performance at higher temperatures, mechanical properties such as tensile and compressive strength, cell-matrix interactions, shape memory retention, and gas permeability. Covalent crosslinking, which is the most widely used crosslinking method [408], can be used with amino acid residues in collagen that contain hydroxyls, amines, and other functional groups. Some of the most commonly used crosslinker molecules are discussed below.
4.2.1. Glutaraldehyde (GA)
Collagen hydrogels have been extensively crosslinked using GA. The aldehyde functional group in GA reacts with hydroxyl or amine groups in collagen and connects the chains via intra- or intermolecular interactions [409–411]. GA forms a tightly crosslinked structure and significantly improves tensile strength and durability of the scaffold while reducing antigenicity of collagen [412]. Being inexpensive, GA is a very commonly used crosslinker. However, the aldehyde functional group of GA causes significant cytotoxicity and inflammation and thus limits its usability. Many detoxifying strategies, such as washing with glycine or citric acid (CA) solutions, have been developed to increase the biocompatibility of GA-crosslinked scaffolds [413,414].
4.2.2. Carbodiimide agents
1-Ethyl-3-(3-dimethyl aminopropyl)-carbodiimide (EDC) is a water-soluble carbodiimide that reacts optimally under acidic pH conditions with an array of functional groups, including carboxyl, hydroxyl, or sulfhydryl [415–417]. EDC reacts optimally in the presence of buffers devoid of any amine or carboxyl groups (e.g., 4-morpholinoethanesulfonic acid) [408,418]. N-Hydroxysuccinimide (NHS) is popularly used in EDC reactions to activate carboxylic acid groups as NHS esters are less susceptible to hydrolysis and improve the efficacy of the crosslinking reaction [419]. Carbodiimide agents are widely used as they are water-soluble and can be removed easily after the reaction. However, the slow crosslinking reaction catalyzed by carbodiimide agents may limit their use in fabricating hydrogels in situ.
4.2.3. Genipin
Genipin (GP) is a natural compound derived by hydrolysis of geniposide from Gardenia jasminoides Ellis fruit. It possesses several functional groups, such as hydroxyls, that can spontaneously react with amino acids and crosslink collagen monomers [412,417,420–422]. GP is biodegradable and has low cytotoxicity [423,424]. It is reported that GP crosslinking may regulate the proliferation, migration, and polarization of immune cells and thereby reduce the immunogenicity of collagens from xenogenic sources [425]. The major disadvantage of GP is high cost, which limits its usage to laboratory-based experimental studies. In addition, GP crosslinking changes the color of the resulting scaffold to blue, which affects light penetration, interferes with colorimetric assessments, and is not preferred aesthetically.
4.2.4. Glycation
Under hyperglycemic conditions, the crosslinking of collagen by glycation naturally occurs in the body [426,427]. In the presence of a non-toxic reducing sugar, such as glucose or ribose, irreversible non-enzymatic crosslinking occurs between aldehyde groups on the reducing sugar and amino groups on collagen. The mechanical properties of collagen gels can be easily altered by glycation in a dose-dependent manner based on ribose addition[428]. However, the major limitation of glycation crosslinking is that it causes ECM dysfunction by altering collagen interactions with cells and other ECM proteins [427].
4.2.5. Citric acid (CA)
CA, a weak, tricarboxylic acid found in citrus fruits, can react with hydroxyl and amine groups of collagen molecules [429,430]. Crosslinking with CA provides additional functionality to the polymer chain because pendant groups on CA can form ester bonds, increase hemocompatibility, balance the hydrophilicity of the polymer network, and enhance the availability of binding sites for bioconjugation [431,432]. It is reported that citrate can induce nucleation of hydroxyapatite in vitro on collagen scaffolds [433]. Crosslinking with CA is advantageous due to its easy availability, low price, and cytocompatibility. However, a major drawback of CA is that high temperatures are required to initiate the crosslinking process, which may denature or alter the polymer structure [430,434].
4.2.6. Other crosslinkers
In addition to the common crosslinkers discussed above, researchers have explored various chemical crosslinkers such as glyoxal [435,436], squaric acid [437], cinnamaldehyde [438], nordihydroguaiaretic acid [439–441], tannic acid [442,443], epoxy-based cross-linkers (e.g., 1,4-Butanediol diglycidyl ether) [444,445], and methyl glycidyl ether and glycerol poly(glycidyl ether) [446] to crosslink collagen hydrogels. Disulfide crosslinking has also been explored to conjugate collagen with poly(ethylene glycol)-di-acrylamide [447]. Disulfide crosslinking will be discussed in detail in Section 4.3.2.1.
4.3. Preparation and crosslinking techniques of HA hydrogels
HA does not natively form physical gels alone and is susceptible to endogeneous degradation. In light of these drawbacks, the hydroxyl and carboxyl reactive groups in HA are often subjected to chemical modifications, crosslinking, and gelling agents to develop HA-based hydrogels with tailored structural, mechanical, and degradation properties while maintaining native biological functions.
4.3.1. HA hydrogels through physical crosslinking
Intermolecular forces, such as hydrogen bonds, hydrophobic/hydrophilic interactions, and ionic/electrostatic interactions, are the basis of physically crosslinked hydrogels [448,449]. Physically crosslinked hydrogels can be tailored to respond to various stimuli such as light, pH, enzymes, temperature change, and fluctuations in redox conditions [450–454]. Physical crosslinking can eliminate the need for extrinsic crosslinking agents and decrease potential cytotoxicity. Physical crosslinking also generally creates hydrogels with less mechanical and chemical stability compared to covalent crosslinking. Since physical interactions are reversible, physically crosslinked hydrogels can be easily created to have self-healing and shear-thinning properties for applications that require extrusion or injections [455,456].
Inclusion complexation is one of the most popular approaches for fabricating physically crosslinked gels. Inclusion complexation involves the interactions of “host” and “guest” molecules, where the “host” has a cavity into which the “guest” compound can be accommodated. The binding strength between the host and guest molecules is attributed to hydrophobic interactions and complementary structural features [457]. One example is a self-assembling hydrogel with β-cyclodextrin-functionalized HA and adamantane-functionalized HA [458]. Similarly, β-cyclodextrin- and azobenzene-functionalized HA resulted in a supramolecular HA hydrogel at physiological conditions with dynamic control of hydrogel properties [459].
Another interesting approach is to functionalize hydrophilic HA with hydrophobic moieties, such as cholesterol, to manifest amphiphilicity, which allows the macromers to self-assemble into hydrogels [460]. Jung et al. reported that hydrophobic interactions between methyl groups of Pluronic F-127 and acetyl groups of HA resulted in enhanced stability of a thermosensitive hydrogel that successfully delivered the drug Piroxicam [461].
Low toxicity and stimuli responsiveness make physically crosslinked hydrogels suitable for drug delivery and encapsulation of living cells. However, physically crosslinked gels can fail to maintain their structural integrity and easily dissolve with changing environmental factors. Therefore, a balance between parameters chosen for fabricating physically crosslinked hydrogels is critical.
4.3.2. HA hydrogels through covalent crosslinking
Monomers can be covalently crosslinked by radiation, enzymes, free radical-generating compounds, polyfunctional compounds, or chemical crosslinkers to develop hydrogels with better mechanical and thermal stability compared to physically crosslinked hydrogels. Several strategies, such as disulfide crosslinking, enzymatic crosslinking, condensation reactions, and click chemistry, are employed to fabricate HA-based hydrogels. However, since most of these strategies change the chemical structure of HA, they can alter its biological activity, such as downstream biochemical cell signaling and the ability of enzymes to degrade HA.
4.3.2.1. Thiolated HA
Thiolated HA is one of the most common modification strategies used to create covalently crosslinked HA hydrogels. Biocompatibility [462], permeation properties [463], and sustained release of a drug [464] from HA-based hydrogels have all been improved through thiolation. HA thiolation is usually performed by chemical reactions between ligands containing free thiol groups and the hydroxyl or carboxylic groups of HA.
Oxidation of thiols by mild oxidizing agents or exposure to oxygen forms disulfide bonds. The use of disulfide bonds in hydrogels is popular for controlled drug release, bioconjugation, and cell encapsulation because cells can cleave these linkages by synthesizing reductants such as glutathione [465]. Disulfide crosslinking is safe, facile, reversible, and allows in situ gelation. However, disulfide formation through thiol oxidation under physiological pH is a very slow process [466]. The rate can be improved by adding oxidants, such as hydrogen peroxide or iodine, or by increasing the reaction pH [467,468]. Bermejo-Velasco et al. adopted an interesting strategy to increase the gelation rate of disulfide crosslinked HA hydrogels by incorporating the electron-withdrawing groups cysteine and N-acetyl-L-cysteine at the β-position of thiol substrates to influence the deprotonation of thiol groups and enhance disulfide bond formation [469].
Thiolated HA can also be crosslinked into gels using crosslinkers such as sodium hypochlorite, carbodiimide, and N-hydroxysuccinimide [470,471]. Crosslinked thiolated HA scaffolds are used in ocular care, wound healing, and tissue engineering [472,473]. For example, thiolated HA was crosslinked via poly(ethylene glycol) PEG diacrylate to form scaffolds for successful delivery of stem cells in vivo [474]. Bian et al. varied the degree of thiol substitution and molecular weight of HA to control gelation and thus developed injectable hydrogels for chondrocyte delivery in vivo [475].
4.3.2.2. Schiff base reaction/dialdehyde HA
Schiff bases are imines prepared by the condensation reaction of a ketone or an aldehyde with primary amines. Dialdehydes can be introduced by oxidizing the hydroxyl groups of HA using sodium periodate [476]. The wide popularity of the Schiff base reaction for fabricating hydrogels is due to its mild reaction conditions, reversibility, fast reaction rate without the usage of toxic crosslinking agents, and its ability to form injectable self-healing hydrogels with in situ gelation [477].
A major drawback of using a Schiff base reaction is that the imine linkages may hydrolyze under acidic conditions. Therefore, in disease conditions, where the pH is usually slightly acidic, these hydrogels would be unstable [478,479]. To address this limitation, Hozumi et al. developed an injectable hydrogel formed from carbohydrazide-modified gelatin and HA monoaldehyde [476]. The hydrogel was cytocompatible, was hydrolytically stable, and supported cell migration in an ex vivo rat aortic-ring assay as compared to other Schiff base crosslinked hydrogels used as controls. [476].
There are many biomedical examples of HA hydrogels crosslinked through the Schiff base reaction. Ma and colleagues used a Schiff base reaction between the aldehyde and hydrazide groups of aldehyde-functionalized HA and hydrazide-modified poly (γ-glutamic acid) to form a self-crosslinking, injectable hydrogel for protein delivery [480]. The hydrogel had tunable gelation time, was cytocompatible, and had favorable mechanical properties. An aldehyde- and adipic acid dihydrazide-functionalized HA hydrogel was crosslinked in situ and was used for gradual release of paclitaxel particles in intraperitoneal cavities [481]. Wang et al. also developed an injectable, biocompatible hydrogel based on dynamic covalent hydrazone bonds between hydrazide-functionalized HA, elastin-like protein (ELP), and aldehyde-functionalized HA [482].
4.3.2.3. Radical polymerization
Radical polymerization is mediated by an initiation source (e.g., temperature, light, redox reaction) that generates radicals to react with functional groups on HA macromers [11]. Photoinitiated polymerization is the most popular radical polymerization technique for creating HA hydrogels. A major advantage of photoinitiated radical crosslinking is temporal and spatial control over hydrogel architecture and properties [483]. For further explanation of temporal and spatial control strategies in hydrogels, see Section 5.3.
Incorporation of methacrylate groups is a very common modification for creating HA-based hydrogels and is achieved by reacting HA with glycidyl methacrylate or methacrylic anhydride through an esterification reaction [484,485]. The methacrylate group can be photocrosslinked using radical initiators such as Irgacure 2959 [486] or riboflavin [487]. The major benefit of methacrylated HA hydrogels is the tunability of hydrogel properties by varying conditions such as the modification degree, monomer concentration, UV exposure time and intensity, and photoinitiator concentration [488,489].
Many studies have taken advantage of the benefits of radical polymerization to form HA hydrogels. For example, methacrylate-functionalized HA scaffolds were used as brain-mimetic ECM models to study the mechanobiology of brain tumor progression [490]. Heparin-HA hydrogels formed by crosslinked methacrylated HA and thiolated heparin supported adipose-derived stem cell spreading, proliferation, and migration in 3D culture [491]. Poldervaart and coworkers developed methacrylated HA hydrogels with suitable properties for 3D bioprinting as well as differentiating MSCs towards the osteogenic lineage [486]. Radical polymerization was explored by Das et al. to develop a novel biocompatible terpolymeric hydrogel using HA, 2-hydroxyethyl acrylate and PEG diacrylate via free radical polymerization for sustained drug release and biomedical applications [492]. The major limitation of radical polymerization is that a fast radical termination reaction is unavoidable and leads to an uncontrolled molecular weight distribution.
4.3.2.4. Crosslinking by condensation reactions
HA macromers can form hydrogels by undergoing condensation reactions. Among the diverse condensation reactions used for HA hydrogel fabrication, esterification through the carboxylic acid and hydroxyl functional group is the most common [466]. Larrañeta et al. fabricated HA hydrogels through esterification between the carboxylic groups of Gantrex S97 and the hydroxyl groups of HA [493]. By conjugating HA with amine-functionalized (−)-Epigallocatechin-3-gallate, Lee et al. developed a bioactive hydrogel for scavenging free radicals and inhibiting the growth rate of cells [494]. Hydrazide-terminated PEG-grafted HA hydrogels were also prepared via condensation reaction by using carbodiimide [495].
A frequently used condensation crosslinking method to fabricate HA-based hydrogels is through covalent ether linkages. For example, the reaction between HA and 1,4-butanediol diglycidyl ether (BDDE) under strongly alkaline conditions forms a stable, covalent ether linkage [496]. Exploring this chemistry, Xue and coworkers recently developed a HA-based hydrogel consisting of a defined ratio of low MW HA to high MW HA crosslinked with varying proportions of BDDE [497]. Divinyl sulfone-based crosslinking also forms covalent ether linkages with HA and the resultant hydrogels are biocompatible and resist enzymatic degradation [484,498,499]. Although hydrogels resistant to enzymatic degradation are beneficial for certain applications such as engineering blood vessels, they may not be suitable for applications where gradual hydrogel degradation is necessary for proper tissue regeneration.
4.3.2.5. Click chemistry crosslinking
Click chemistry describes a family of reactions that typically have a high thermodynamic driving force and proceed under mild conditions, generate high yields, and produce limited and easily purified byproducts [500,501]. Diels-Alder (DA) and Huisgen azide-alkyne cycloaddition reactions are two types of click chemistry used in developing HA hydrogels [466,479].
A DA click chemistry reaction is a selective [4 + 2] cycloaddition between a diene and a dienophile in the absence of a catalyst. The reaction is highly efficient and proceeds under physiological conditions [502,503]. Common Diels-Alder HA crosslinking involves furan (diene) and maleimide (dienophile) reactions. Both maleimide-functionalized HA and furan-functionalized PEG [503], and furan-functionalized HA and dimaleimide PEG has also been utilized to manufacture hydrogels [502,504]. The gels have been studied for biopharmaceutical delivery applications, cartilage tissue engineering [504], and neurite outgrowth [505]. Other furan derivatives, such as the more electron-rich methylfuran, have been appended to HA and result in faster gelation times (Fig. 5) [506]. The use of horseradish peroxidase (HRP) prior to DA chemistry enabled fast gelation of the material, and the resulting injectable hydrogels had shape memory and anti-fatigue properties [507]. Similarly, dual-crosslinking has been demonstrated by photo-crosslinking of HA-Furan with lithium phenyl-2,4,6-trimethylbenzoylphosphinate followed by a DA reaction [508]. A recent study introduced inverse electron-demand DA (IEDDA), which is a reaction involving electron-deficient diene and electron-rich dienophile, the opposite of DA [509]. IEDDA was utilized to crosslink HA–norbornene and HA–methylphenyltetrazine and resulted in gels with a unique ability to tune gelation time independently of stiffness [510]. Other examples of HA DA chemistries include post-functionalization of crosslinked HA-PEG hydrogels with cyclohexene derivative groups using photoinduced radical thiol−ene reactions [511].
Fig. 5.
Replacing furan with methylfuran accelerates the Diels-Alder (DA) reaction. (A) Covalently cross-linked hydrogels can be fabricated using furan-maleimide DA chemistry. (B) The methylfuran-maleimide DA reaction proceeds faster than furan-maleimide at pH 7.4. Reprinted with permission from L.J. Smith, S.M. Taimoory, R.Y. Tam, A.E.G. Baker, N. Binth Mohammad, J.F. Trant, M.S. Shoichet, Biomacromolecules 19 (2018) 926–935. Copyright 2018 American Chemical Society [506].
Huisgen-type copper-catalyzed azide-alkyne cycloaddition (CuAAC) has been used for the crosslinking of HA gels This reaction occurs through a 1,3-dipolar cycloaddition of azides and terminal alkynes [500,501,512] and typically occurs in the presence of a selective Cu(I) catalyst [513]. The first application of the Huisgen reaction to create HA gels involved 11-azido-3,6,9-trioxaundecan-1-amine-HA and propargylamine-HA and resulted in fast gelation (120–130 seconds) [501].
Although the copper catalyst is selective and thus a minimal amount of catalyst is required to produce biomaterials, there are concerns about copper toxicity. Thus, strategies to perform the Huisgen addition in copper-free environments have been developed. Strain-promoted azide-alkyne cycloaddition between cyclooctyne and azide groups [514] avoids copper and has been used with azide-modified HA and cyclooctyne-modified HA undergoing cycloaddition at physiological conditions [515]. Another example is HA-PEG4-DBCO (dibenzocyclooctyl) crosslinked with a 4-arm PEG azide [516] in a copper-free environment. Removing or limiting copper catalyst from the reaction enables biocompatible scaffold formation.
4.3.2.6. Horseradish peroxidase (HRP) Enzymatic Crosslinking
Enzymatic crosslinking of HA is an attractive option for hydrogel fabrication due to fast and controlled gelation [517], tunable mechanical properties [518], and mild, physiological reaction conditions [479,504]. These desirable properties lead to multiple applications for the gels, including cartilage tissue engineering [504,519,520], neural development [521], drug and protein delivery [517,518,522], injectable bone cement [523], and expansion of human embryonic stem cells [524].
HRP has been used to catalyze the crosslinking of aromatic rings in phenolic hydroxyl groups through C-C and C-O coupling in the presence of H2O2 [525]. This reaction was used to crosslink tyramine-modified HA (HA–Tyr) and form hydrogels [524]. Additives, such as silk [526] and gelatin [527], can be used to improve the mechanical integrity and add resistance to degradation. Other enzymatically crosslinked gels include an injectable composite gel developed from tyramine-modified alginic acid and tyramine-substituted HA [528], and HA grafted with a dextran–tyramine conjugate [519,520].
Despite its promise in various applications, pure HRP is expensive to produce and results in low yields [529]. Consequently, other catalysts, including hematin [530,531] and an enzyme-mimicking chitosan-g-hem biocatalyst [532], have been studied to replace HRP.
4.4. ColHA hydrogels
Natural ECM mechanical structure is important in regulating cell behaviors [533,534], and hydrogels composed of both collagen and HA can combine the features of these two macromolecules to better mimic ECM structure and function. The fabrication methods used to prepare ColHA hydrogels determine their biophysical properties, such as the interior microstructure, mechanical strength, swelling properties, and transport of molecules. Therefore, these blended hydrogels can provide insight into the relationship between ECM composition and the biophysical properties of the gel. Various hydrogels built from combining collagen and HA have been developed for biomedical applications, including drug delivery, cell culture, tissue regeneration, and wound healing.
Based on the crosslinking states of collagen and HA during the formation of the hydrogel network, there are three main types of ColHA hydrogels: a semi-interpenetrating network in which only collagen is crosslinked, a semi-interpenetrating or interpenetrating network in which HA is chemically crosslinked, and a crosslinked copolymer network in which collagen and HA crosslink with each other. We will discuss these types of ColHA networks below.
4.4.1. ColHA semi-interpenetrating network in which only collagen is crosslinked
Collagen can self-assemble into fibrillar structures to form a hydrogel network without the addition of chemical crosslinkers. HA can be added to the collagen solution before polymerization and is retained in the fibrillar collagen network. For instance, hydrogels consisting of high MW HA (1.5−1.8 MDa) and a blend of collagen I and II demonstrated that HA did not inhibit gel formation and that HA incorporation percentages were similar across different collagen I/II ratios [535]. Another example mixed low MW HA (150–300 kDa) with collagen to form a semi-interpenetrating network, and gelatin microspheres with growth factor were loaded into the hydrogel [536]. The composite hydrogel system supported the growth and chondrogenic differentiation of MSCs.
Many factors, including HA MW and the modification of HA, can be used to modulate ColHA hydrogel properties without crosslinking agents. Xin and coworkers found that incorporation of low MW HA (155 kDa) increased the elastic modulus of ColHA gels, whereas no improvement was obtained using high MW HA (1.2 MDa) [537]. It was observed that low MW HA was interspersed within the collagen network and completely coated the fibrils. These observations may be explained by higher chain mobility and weaker homologous interactions, which may have led to stronger interactions with collagen. In a recent study, adding low MW HA (~400 kDa) increased stiffness, mean pore size, and mean fiber radius of the collagen matrices, but the transport parameters were not affected [538]. The increase in pore size may be attributed to HA-induced swelling [539], whereas HA associating around the collagen fibers in the process of collagen fibrillogenesis may cause an increase in fiber radius [537,538]. HA grafted with a collagen-binding peptide was also used to prepare a hydrogel with collagen, and the storage modulus of this hydrogel was more than one order of magnitude higher than the control ColHA hydrogel [540].
Many ColHA hydrogels have been fabricated by introducing HA polymers into covalently crosslinked collagen frameworks. For example, PEG ether tetrasuccinimidyl glutarate (4SPEG) was reacted with amino groups in collagen to form a stable semi-interpenetrating network hydrogel for chondrocyte delivery [541]. The study used mild reaction conditions, which enabled the incorporation of bioactive factors and cell encapsulation during gel formation. The same crosslinking method was used in another study for ocular drug delivery [542]. Another example used riboflavin to photo-crosslink collagen (Fig. 6) [543]. The hydrogel was stiffer and had delayed enzyme-triggered degradation compared to a physically crosslinked collagen hydrogel. To retain HA in the hydrogel network, HA was pre-crosslinked and ground into a powder to create a dispersion of small HA beads in the collagen hydrogel network. Less than 5% of the HA beaded structures released from the hydrogel after 6 days compared to ~31% of normal HA. In summary, covalent crosslinking of collagen can form stable networks with adjustable mechanical properties and degradation rates, and the incorporation of HA in different forms improves the biomedical application of these hydrogels.
Fig. 6.
Schematic representation of (A) photo-crosslinking of collagen with riboflavin, (B) fabrication of a ColHA composite hydrogel. Reprinted by permission from Springer Nature Customer Service Centre GmbH: Springer Nature Drug Delivery and Translational Research Riboflavin-induced photo-crosslinking of collagen hydrogel and its application in meniscus tissue engineering, J. Heo, R.H. Koh, W. Shim, H.D. Kim, H.-G. Yim and N.S. Hwang, Copyright 2015 [543].
4.4.2. ColHA semi-interpenetrating and interpenetrating networks in which HA is chemically crosslinked
HA can form hydrogels when HA chains are chemically modified and covalently crosslinked. Numerous HA hydrogels with different crosslinking methods and specific properties have been developed as discussed in Section 4.3. Collagen is a relatively rigid polymer compared to HA [544], and therefore embedding collagen into a crosslinked HA network can result in considerable changes to the physical and biological properties of the resultant composite hydrogel. Collagen I molecules can assemble into fibrils at concentrations as low as 4.73 μg/mL at 29 °C with the critical concentration decreasing as temperature was increased to 41°C [545]. The concentrations of collagen in ColHA gels discussed in this section are much higher than these critical concentrations. Collagen in a crosslinked HA network can change the interactions between cells and HA hydrogels even at low concentrations.
Adding collagen to crosslinked HA networks improved cell behavior and tissue regeneration. For example, 0.5 w/v% collagen was added to HA hydrogels, which were formed by photo-crosslinking of methacrylated HA [546]. GAG and proteoglycan production in in vivo cartilage defects improved respectively compared to hydrogels made only of HA. Collagen was added at 0.2% to acrylated HA hydrogels crosslinked by thiol groups that contained MMP-sensitive peptides and improved the adhesion and migration of ECs [547]. Chaudhuri et al. developed a ColHA interpenetrating hydrogel with collagen at 2.5 mg/mL and HA crosslinked by dynamic hydrozone bonds, which contributed to stress relaxation [548]. The collagen network provided the fibrillar architecture and mimicked the fiber realignment process during cell spreading in ECM.
Collagen concentration also affects the properties of crosslinked HA gels. Increasing the ratio of collagen to a modified PEG diglycidyl ether crosslinked HA (MHA) increased tensile strength, water uptake, and chondrocyte viability but decreased porosity of the hydrogel [549]. In another study, hybrid scaffolds were prepared by adding 0.1, 0.3, or 0.5% collagen to an ethylene glycol diglycidyl crosslinked 1% HA hydrogel [550]. The scaffolds showed an increase in tensile strength and prolonged degradation time in vitro with increasing collagen concentration.
4.4.3. Collagen and HA co-crosslinked hydrogels
In addition to forming semi-interpenetrating or interpenetrating networks with collagen and HA, HA and collagen chains can chemically crosslink with each other to form stabilized hydrogel networks. A common example is the EDC- and NHS-catalyzed crosslinking reaction between a carboxyl group in HA and a primary amine in collagen [551–553]. Calderon et al. showed that increasing the EDC/NHS concentration from 0 to 48 mM significantly decreased the swelling ratios of collagen II and HA hydrogels whereas compressive stress dramatically increased [551]. The highest EDC concentration (48 mM) resulted in hydrogels with the highest elastic modulus and the most resistance to degradation, but cells loaded in hydrogels made with a lower EDC concentration (8 mM) had a higher proliferation rate.
Diglycidyl ether is another common crosslinking agent that reacts with both the hydroxyl groups in HA and the amino groups in collagen. For instance, PEG diglycidyl ether was added to a solution of collagen and HA to form a crosslinked network [554]. As the HA and collagen concentrations increased, the water uptake percentage increased due to the hydrophilicity of HA. On the other hand, interior pore sizes and degradation rates decreased because of the more ordered and higher concentration of polymer chains. When prednisolone was loaded into the hydrogel, increasing the concentration of HA and collagen reduced the rate of drug release due to the smaller pore sizes.
Other crosslinking techniques such as photo irradiation and enzyme catalysis can be applied to initiate the crosslinking of HA and collagen polymer chains that have been modified with methacrylate, phenol groups, or other moieties capable of free-radical polymerization [555,556]. Ying and co-workers modified collagen and HA with phenol groups and formed a hydrogel when mixed in the presence of HRP and H2O2 as shown in Fig. 7 [555]. The gelation time ranged from several seconds to minutes depending on the concentration of HRP and H2O2. The hybrid hydrogel enhanced in vitro cell proliferation compared to hydrogels containing only collagen. Moreover, the hybrid hydrogel induced only a mild inflammatory response and promoted full-thickness wound healing.
Fig. 7.
ColHA hydrogel prepared via HRP-catalyzed crosslinking of collagen and HA modified with phenol groups. Cells can be loaded into the hydrogel for 3D culture, and the hydrogel can be used for wound healing [555]. Reprinted from Materials Science and Engineering: C 101, H. Ying, J. Zhou, M. Wang, D. Su, Q. Ma, G. Lv, and J. Chen, In situ formed collagen-hyaluronic acid hydrogel as biomimetic dressing for promoting spontaneous wound healing, 487–498, copyright (2019), with permission from Elsevier.
Covalent crosslinking of HA and collagen together can change the properties of composite hydrogels in a wide range, and further work needs to be performed to identify optimal polymer and crosslinker concentrations for applications in cell culture, drug delivery, and tissue repair.
4.4.4. Summary of ColHA hydrogels
ColHA hydrogels combine the biological properties of these two ECM components and have wide applications in biomedical fields. The concentrations, chemical modifications, crosslinking types, and fabrication techniques are the main factors that affect properties such as the mechanical strength, internal microstructure, swelling behavior, and degradation profiles. When using uncrosslinked HA in ColHA hydrogels, HA-collagen interactions and HA retention in hydrogel networks strongly depend on the HA MW and concentration. HA crosslinking can increase the mechanical strength and slow down degradation of ColHA hydrogels, but HA bioactivity may change due to functional modification and crosslinking. Literature reports that lower levels of HA functionalization promoted greater neurite growth as compared to higher levels of functionalization [557].
A variety of other polymers, such as other ECM components (e.g., laminin [558–560] and chondroitin sulfate [561–563]), synthetic polymers (e.g., poly(ε-caprolactone [564] and poly(vinyl alcohol) [565,566]), and natural polymers (e.g., chitosan [567,568] and alginate [569]), can also be used as additives to ColHA hydrogels to meet specific application requirements. The fabrication of ColHA hydrogels with other additives is beyond the scope of this review.
5. Modulating Properties of Collagen and HA hydrogels
Collagen fibrillogenesis and polymerization form hydrogel networks with characteristic microstructure, which impacts the functional properties of hydrogels. In ColHA hydrogels, HA influences the microstructure and mechanics based on its association with collagen. This section discusses the regulation of internal fibril microstructure and mechanical properties of collagen and/or HA hydrogels. Physical processing methods can also be utilized to create hydrogels with the desired structures and properties. This section will focus on molding, fluidic devices, and three dimensional bioprinting methods. Finally, to mimic the dynamic properties of ECM, the fabrication of collagen and/or HA hydrogels with temporal and spatial controlled properties will be discussed.
5.1. Internal microstructure and mechanical properties
Collagen molecules aggregate to form fibrils that further develop into fibril bundles and fibers to create a 3D mesh. The configuration of this collagen network is broadly known as the microstructure. Within collagenous tissues and hydrogels, characteristic microstructural features include fibril density, diameter, length, alignment (orientation), pore size, and degree of crosslinking, among others (Fig. 8) [392]. These features are part of a quintessential biological relationship in which the structure of the tissue dictates functional properties such as mechanics, molecular transport, and cellular behavior [392,570–573]. By varying experimental factors such as collagen source, concentration, solubilization method, and crosslinking technique, the diameter of fibrils can range from 0.05 to 5 μm and pore spaces vary from one to hundreds of microns in diameter [243,245,574–576]. Researchers can use this understanding of the factors and mechanisms that determine the microstructure to engineer desired properties for biomedical applications.
Fig. 8.
(A) Hierarchical structure of collagen demonstrating the collagen molecule (M), fibril (F), and proteoglycan (PG) structures subject to variable deformation, tendon strain (εT), fibril strain (εF), and molecule strain (εM) for different structural levels. Reprinted from Progress in Material Science, Vol. 52. P. Fratzl, R. Weinkamer, Nature’s hierarchical materials, 1263–1334, 2007, with permission from Elsevier [577]. (B) Confocal reflectance microscopy image of representative collagen microstructure metrics including pore size (P), fibril diameter (D), and orientation (θ). Scale bar = 20 μm.
To fully characterize hydrogels, changes in microstructural environment and their impact on functional properties should be observed. Collagen and HA hydrogels are viscoelastic, which means that they both store energy elastically and viscously dissipate energy in a time-dependent manner in response to an applied deformation [578,579]. Physiologically, this viscoelasticity is one reason that collagenous tissues, including muscle, bone, tendon, and skin, are so ubiquitous and versatile. Energy storage and transmission occur with the loading of muscle, tendon, and bone and result in locomotion, whereas these same tissues dissipate energy to constrain motion and protect the body from wear due to repeated loadings and high stresses [13,580–583].
The viscoelastic properties of collagen are due to its ability to maintain loads upon deformation and dissipate this stored energy via reconfiguration on multiple levels of its hierarchical structure [584–586]. The fluid component contained within the interfibrillar space also plays a role in collagen reorganization [587]. For collagen hydrogels with HA incorporated, changes to fibrillogenesis and the properties of the interstitial fluid likely alter the viscoelasticity of the system [588]. Given that HA facilitates changes to collagen hydrogels, an understanding of the microstructure and mechanics of in vitro ColHA hydrogels is important for the development of these materials for unique biological applications.
5.1.1. Collagen microstructure and mechanical properties
For collagen hydrogels, manipulating parameters such as pH, collagen self-assembly temperature, ionic strength, and collagen concentration has been widely reported to modulate collagen microstructure, and these effects are summarized in Section 4.1. Collagen source and age variation are discussed in Section 3.1.1. Additional factors including magnetic fields, ultrasonication, and biomolecular composition have also been shown to control aspects such as collagen alignment, fibril size, and pore size [170,589].
Mechanistically, changes in collagen fibril length and diameter stem from polymerization of collagen monomers and the fusion of distinct collagen fibrils [590]. Garvin et al. demonstrated the use of ultrasound to localize higher temperatures within a 3D collagen matrix and control fibril diameter [591]. The elevated temperatures decreased the duration of fibril nucleation, prevented lateral aggregation, and resulted in thinner fibrils. Thinner fibrils may lead to increased resistance to creep deformation, which is defined by changes in strain under a small constant stress, due to maximizing the surface area of small fibers to distribute shear stress [592]. The ability to resist creep deformation imparts collagen hydrogels the ability to maintain their shapes over time and may be important for studies of longer duration such as extended cell culture. On the other hand, thicker fibers may impart greater collagen tensile strength due to increased intrafibrillar covalent crosslinking [592]. Conversely, Roeder et al. demonstrated a higher importance of fibril length rather than diameter in dictating tissue stiffness [593]. The importance of fibril length is perhaps related to the ability of longer fibrils to undergo additional triple helix internal stretching, the axial sliding of individual triple helices past each other, or the unfolding of the crimp regions to alleviate tissue strain [583,594,595].
In addition to fibril shape, collagen fibril orientation plays a unique role in hydrogel mechanics and has further relevance in 3D tissue culture. Homogenous orientation of collagen fibrils in the axial direction lends tissues, such as tendons, outstanding tensile strength [596], particularly in the axial direction of aligned fibers, since the forces resisting deformation are also aligned. Upon mechanical deformation of the aligned collagen matrices, resistance to tensile strain is conferred at the sub-fibrillar level due to interfibrillar covalent crosslinks and the intrinsic stiffness of individual collagen molecules in the axial, rather than transverse, direction. Furthermore, fibrils sliding relative to each other in the direction of deformation impart extensibility, which is the ability to stretch without fracture under high stress applied to the aligned matrices [583,597]. Manipulating alignment in vitro can be useful for controlling hydrogel mechanical properties [598]. Ng et al. demonstrated the creation of aligned collagen networks as a downstream result of interstitial fluid flow-imposed cellular alignment of the ECM [599]. Another approach utilized small mechanically-induced strains to introduce similar collagen fibril reorganization [600,601]. Other methods for the creation of aligned collagen matrices include magnetic particle manipulation [602], shear flow of the pre-polymerized solution [603], and, for radial alignment akin to tumor or other unique ECMs, rotational force applied via acupuncture needle [604,605].
Finally, the pore space within the collagen matrix is a microstructural characteristic relevant to mechanical integrity, as well as cellular and molecular transport within collagen hydrogels. By using a two temperature polymerization process (first incubating at 22, 27, or 32 °C for different durations and then at 37 °C to finish polymerization), Yang and coworkers demonstrated that temperature can be utilized to create variable pore sizes [397]. Scaffolds polymerized at lower temperatures had a longer gelation lag phase, and the result was larger pores and a less dense collagen network. One potential explanation is that an increased lag phase duration may allow more time for the formation of thicker fibers via lateral aggregation of monomers. Yang and coworkers also demonstrated that collagen scaffolds polymerized at lower temperatures, which had increased pore sizes, had lower equilibrium storage moduli. Furthermore, although cellular motility is often correlated with matrix stiffness [606,607], glioma invasive distance was observed to be independent of modulus and dependent on collagen hydrogel pore size and fiber thickness [397].
In vivo, the properties of collagen biopolymer networks are the result of the physical, chemical, and biological environment, and replicating these conditions in vitro is therefore useful as a means to study the mechanisms that drive each unique presentation. Thermodynamic and kinetic changes in collagen fibrillogenesis affect pore size, fiber dimension, and orientation, and these parameters dictate tissue properties and ultimate cellular behavior. As a composite biomaterial, however, ColHA matrices have a different biochemical environment through the inclusion of HA, and it is worth investigating how this may alter the fibrillogenesis process.
5.1.2. Influence of HA on microstructure and mechanical properties of collagen
The hydrophilicity, charge density, and MW variability of HA may have a direct impact on the resultant hydrogel microstructure and mechanics based on its association with collagen. Emphasis has been placed on small leucine-rich proteoglycans, due to known protein core interactions with collagen and their ability to influence collagen architecture in vivo, and other GAGs, but HA may play a role in physiology via processes not dependent on similar interactions [608–611]. The effects of HA on collagen microstructure likely stem from electrostatic, hydrophilic, and ionic interactions and depend on HA molecular weight, and subsequently the resulting ColHA hydrogel properties strongly depend on the specific formulation. HA without chemical modifications can act as crowding regent, diffusional regulator, water absorbance agent, and viscosity regulator to affect collagen fibrillogenesis and properties of collagen gels.
The following studies report the effects of HA concentration on collagen pore size; however, their results yield opposite conclusions. Specifically, they do not account for confounding parameters such as collagen type (pepsin vs acid solubilized) and treatment (polymerization conditions) and how these may yield different overall scaffold properties. For example, Entekhabi and coworkers created a lyophilized collagen and HA matrix in the various collagen:HA weight ratios of 100:0, 98:2, 95:5, and 90:10 [612]. Increasing HA ratio increased water uptake and caused the average pore size to increase from 37.29 ± 12 μm in the 100:0 gels to 180.35 ± 38 μm in the 90:10. However, there was no significant impact on compressive moduli. Yang et al. observed that the incorporation of HA percentages >2% within collagen gels decreased the pore size of the network compared to pure collagen due to the apparent increase in fibril number and decrease in fibril size [613]. Additionally, the decreased pore size led to an associated increase in modulus. Whereas the lyophilized scaffolds from Entekhabi et al. demonstrated changes in pore size due to HA-induced water uptake after scaffold formation, the pore size differences demonstrated by Yang et al. resulted from HA interactions with collagen during the polymerization process. Importantly, they compared pepsin-treated and acid-solubilized collagen in the presence of HA. They observed more dramatic changes for the telopeptide-containing acid-solubilized collagen, and these results indicate that the non-helical ends of collagen may be crucial for HA influence on collagen structure. Both studies demonstrate effects of HA on collagen microstructure and mechanics and show that both an increase and decrease in pore size is possible with HA inclusion. These examples emphasize the variability of ColHA hydrogel systems and the caution needed when making comparisons across studies. Lack of standardization between studies may be misleading to readers at first; however, these experiments increase the overall body of knowledge within the field by showing how different material or fabrication parameters alter the resulting ColHA scaffolds.
Altering the viscoelasticity of ColHA hydrogels can adjust the kinetics of the fibril forming process and therefore alter the microstructure [14]. Tsai and coworkers found that increasing alginate concentration increased polymerization lag time, slowed total assembly time, and increased fiber diameter whereas increasing HA concentration had the opposite effect, despite both polysaccharides being negatively charged. Increased pre-polymerized solution viscosity at high concentrations of either alginate or HA could both potentially lead to limited collagen molecule mobility and decreased intermolecular interactions, resulting in slow polymerization and thicker fibrils. However, the greater water-drawing capacity of HA compared to alginate could reverse this effect. With its extremely hydrophilic nature, HA may draw more water away from collagen within the pre-polymerized solution and lead to increased local concentrations of collagen molecules, greater intermolecular interactions, quicker polymerization, and thinner fibrils. Indeed, their study showed shorter lag phase, assembly time, and smaller fiber diameters in HA-containing gels. A similar reduction of fibril diameter and increase in fibrillogenesis rates was demonstrated with CS, and the observation was hypothesized to be a result of either an increased number of collagen nucleation sites or a change in the nucleation site shape from electrostatic charge [614].
Kreger and Voytik-Harbin observed that adding 0, 0.5, and 1 mg/mL HA to collagen hydrogels had negligible effects on fiber diameter but still resulted in a decrease in mechanical strength. For example, the storage modulus lowered from 24.3 ± 5.2 Pa in pure collagen to 15.8 ± 3.4 Pa in collagen containing 1 mg/mL HA [615]. In other words, the addition of HA altered the viscoelasticity but not the microstructure. These results directly contrast with the study by Tsai et al [14]. These findings suggest that HA did not lower the modulus of collagen through alterations to the fibers but rather through processes such as swelling or HA-mediated sliding of fibrils within the interstitial space. However, these parameters were not directly measured. Additional studies that capture all relevant measurements including pore size, fiber length, fiber diameter, swelling, and storage and loss modulus should be performed under controlled conditions to fully understand the impact of HA on fibrillogenesis.
Chemical modification and alternate fabrication techniques of ColHA hydrogels impart additional control over collagen microstructure that do not necessarily rely on fibrillogenesis. For instance, collagen gels containing increasing amounts of thiol-modified HA (30–100% v/v) increased the porous nature of the scaffolds, provided a stable microenvironment for the prevention of cell-induced matrix contraction, and resulted in better cell proliferation and clinical flexibility compared to either pure collagen or pure thiolated HA hydrogels [616]. Blend hydrogels made with higher thiolated HA volumes and less collagen led to decreasing storage and loss moduli, and these softer gels may be better suited for clinical use due to lower forces needed for injection. Similarly, Wang et al. combined collagen I and II with HA (compositions of 0, 33, 50, and 67%) using a carbodiimide crosslinker to produce freeze dried hydrogels [617]. Increasing HA incorporation led to increasing pore sizes ranging from approximately 75 μm to 200 μm in diameter. The compressive modulus of the collagen scaffold with 67% HA had a reduced compressive modulus (1–2 kPa) compared to the pure collagen I scaffold (6.3 kPa), and this result was attributed to decreased density and mechanical strength of the struts composing the mesh network. It is important to note that the structural characteristics of these scaffolds did not depend on collagen fibrillogenesis but rather on the freeze-drying preparation and carbodiimide crosslinking.
5.1.3. Summary of relationship between microstructure and mechanical properties
As discussed, HA-induced changes to collagen network formation are variable for different formulations. In general, however, it is known that HA elicits some control over the ultimate properties of collagen hydrogels through interactions with collagen before or during fibrillogenesis that result in different fiber network formation and/or through alterations in the viscoelasticity of bulk gels after polymerization. Additional studies, with careful comparison of measurement conditions such as strain and frequency, are required to fully understand the cause-and-effect relationship of these biopolymers.
5.2. Processing methods for collagen and HA hydrogels
The numerous chemical modifications for collagen and HA hydrogels lend these materials flexibility in physical processing methods, such as molding and bioprinting, for the formation of hydrogels useful in biomedical applications. Physical processing of these materials can be utilized to create hydrogels with a desired structural or functional shape on macro-, micro-, and nanoscopic scales. An analysis of these methods is helpful to understand the purpose of each processing technique with the overall goal of developing a better understanding of collagen- and HA-based biomedical technologies.
5.2.1. Molding and fluidic devices
Various high throughput methods of creating microgels, which are micron-sized hydrogel particles, of consistent size have been developed [618]. Microgel fabrication is often achieved using a biphasic system; a hydrophilic phase carries the hydrogel prepolymer solution, and a hydrophobic phase shapes the aqueous phase prior to polymerization. Several phase-based strategies have been developed to shape the aqueous prepolymer solution. Thomas and coworkers employed a solid phase strategy using a hydrophobic surface (e.g., Teflon-coated tape) to form a microgel using the surface tension of the aqueous prepolymer solution [619]. For more control over the form of the microgels, Yeh et al developed micro-molds made of poly(dimethylsiloxane) (PDMS) to cast microgels with well-defined geometries [620].
Liquid-phase fabrication strategies involve mixing an organic solvent around droplets of the aqueous prepolymer solution. Some groups slowly dripped the aqueous phase into the organic phase while stirring [621,622], whereas others dispersed the aqueous phase throughout the organic phase through vortexing [623], mechanical homogenization [622,624], or sonication [616]. For greater control of the fabrication process parameters, custom microfluidic chambers have been developed [618,625–627]. Channels and compartments etched into PDMS using soft lithography can create pathways for the flow of the aqueous and organic phases. The intersecting flows of these two phases create droplets of the prepolymer phase. The shape of the droplet is controlled by the flow rate of the phases and the microfluidic geometries [626].
Using these molding and microfluidic techniques, researchers have developed collagen and HA microgels for drug delivery and tissue engineering applications. Due to their small size when dried, HA microgels suspended in oil have been employed as transdermal drug delivery vehicles, which swell and release their payload once inside the skin [622]. Researchers can use microgels for high-throughput screening of cell scaffold parameters. For example, Fontana et al. made collagen II and HA microgels of varying concentrations and stiffnesses to determine their effect on encapsulated adipose-derived stem cells [628]. Sideris et al. fabricated large cell scaffolds by annealing individual HA microgels together to form a microporous bulk material (Fig. 9) [627]. Furthermore, due to their ease of injectability resulting from their granular form, suspensions of HA microgels can be used as bioinks for 3D printing of hydrogel structures [629].
Fig. 9.
Porous bulk hydrogel made from annealed microgels support fibroblast proliferation. HA microgels labeled green with Alexa-Fluor 488, and human dermal fibroblast actin and nuclei stained with rhodamine-B conjugate of phalloidin (red) and DAPI (blue) respectively. Adapted with permission from E. Sideris, D.R. Griffin, Y. Ding, S. Li, W.M. Weaver, D. Di Carlo, T. Hsiai, T. Segura, ACS Biomater. Sci. Eng. 2 (2016) 2034–2041 [627]. Copyright 2016 American Chemical Society.
5.2.2. Three dimensional bioprinting (3DBP)
Extrusion bioprinting is a method of depositing fluid and/or gel-like material through a nozzle into desired geometries on a flat platform. After deposition, these materials typically retain their structure through thermal, physical, and/or covalent crosslinking that confers stable mechanical properties. The extrusion bioprinting method is a flexible bioprinting technique due to its compatibility with a wide range of materials and its ability to print constructs with physiological cell densities [630,631].
Light-based bioprinting (stereolithography and digital light processing) utilizes ultra-violet (UV) or visible light to polymerize photocrosslinkable materials such as methacrylated collagen and HA to form structures in situ. Compared to extrusion bioprinting, light-based techniques have higher resolution, or a smaller minimum feature size, which is governed by the cross-sectional area of a light beam rather than a clog-prone extrusion nozzle [632]. Although the high resolution of this method is advantageous, light-based bioprinting is limited to solely photoreactive materials, requiring chemical modifications for utility with collagen and hyaluronic acid, and has potential to impact viability of cell-laden constructs through UV exposure [633].
3D electrohydrodynamic jetting, developed from electrospinning, is a non-traditional method of bioprinting that uses electric potential to draw polymers from a thin needle and deposit them on a surface in a controlled fashion [634,635]. Much like light-based methods, this technique excels in precision and resolution, but the process is less cell compatible due to high electric potentials and the use of harsh solvents. Additional printing methods include droplet-based techniques, such as inkjet and laser-assisted bioprinting, which utilize force to deposit picoliter volumes of material in a controlled fashion [636,637]. These printing techniques have limited utility for unmodified collagen and HA bioinks due to the incompatibility of inkjet printing with highly viscous materials.
Overall, these 3DBP techniques enable different architectural possibilities for the fabrication of collagen and HA tissue matrices. Simplified schematics of each of these 3DBP techniques are depicted in Fig. 10.
Fig. 10.
Simplified working principles of common 3DBP techniques. Blue represents bioink. (A) Extrusion printing involves deposition of material via piston, pneumatic, or screw syringe such that the material is well-supported and maintains its shape. (B) Inkjet printing, and droplet printing in general, deposits small volumes of material that flattens as it is printed. Another option is to closely print drops that then coalesce into lines rather than stay as distinct drops. (C) Electrodynamic jetting and (D) electrospray utilize a voltage (V) to extrude fibers using a charged focusing ring (orange) or random dispersion, respectively. Finally, (E) stereolithography uses light to crosslink photoactive material on a print surface (grey) using a mirror-directed laser whereas (F) digital light projection crosslinks the entire plane simultaneously.
As 3DBP is an emerging field, current literature boasts many novel tissue engineering applications involving collagen, HA, and ColHA gels. Many researchers make use of collagen and HA separately in bioinks for extrusion-based bioprinting and may often combine these materials with additional stabilizing components or chemistries to facilitate printing. Since an optimal bioink should retain characteristics of native ECM, one study printed solutions of pure, high concentration collagen [638]. Although pure collagen provides a high density of cell-binding domains for cellular compatibility, viscous collagen dispersions may be amenable to nozzle clogging. As such, Moncal and coworkers utilized pluronic F127-doped collagen to thermally modulate extrusion kinetics for optimal printability [639]. For supplemented collagen dispersions, printability is determined by the collagen self-assembly kinetics as well as the mechanical properties of the supplement material, whereas typical HA bioinks are controlled via chemical crosslinking methods. Many researchers utilize acrylated and methacrylated HA to photocrosslink printed material for high resolution scaffolds [486,640,641].
Hybrid collagen and HA bioink designs facilitate the combination of these two distinct ECM components using advantageous chemistries that enhance printability as well as physiological relevance. For example, Mazzocchi et al. utilized methacrylated collagen and thiolated HA in a 3:1 ratio to develop a bioprinted liver model with optimal printability and cellular compatibility [642]. This technique allows sufficient shape retention of the scaffold and enables post-print photocrosslinking to dynamically modulate hydrogel mechanical and swelling properties. Furthermore, Fisher and coworkers used electrospinning without the use of chemical modification to make hybrid nanofiber mesh scaffolds for bone tissue engineering [643]. A distinct advantage of this method is the nano-scale fiber resolution. Nano-scale structures are desirable for bone tissue models since porous morphological characteristics result in high surface area that enable scaffold interaction with cells as well as the addition of a high density of bioactive molecules [644,645].
The application of 3DBP toward the fabrication of collagen and HA hydrogels has been extensively explored. Although these materials present some processing challenges due to viscosity limitations, temperature requirements, and the potential need for chemical derivatization, 3DBP techniques have been successfully used to create several unique hydrogels tailored for numerous applications. In applications where 3DBP is inadequate to address experimental needs, a variety of other processing techniques, such as lyophilization and plastic compression, may be utilized to create unique hydrogel microstructures but at the cost of control over scaffold geometry. This limitation may spur on the development of novel 3DBP techniques that address the processing challenges inherent for these materials and thus lead to the possibility of creating scaffolds for probing specific microstructure-dependent biological phenomena such as cancer invasion, metastasis, and dormancy; tissue regeneration; and drug transport and delivery.
5.2.3. Other techniques
In addition to traditional processing methods, there are a host of techniques that are suited to manipulate the structures and properties of collagen and HA hydrogels. Freeze drying, or lyophilization, is a processing technique to prepare solid collagen scaffolds of controlled porous structure [10]. For tissues that display a consistent porous nature such as adipose tissue, Davidenko and colleagues prepared composite ColHA scaffolds by controlled freeze drying and crosslinking with EDC [646]. These scaffold systems demonstrated an increasing elastic modulus, swelling potential, and resistance to dissolution with increasing amounts of HA. Controlled freeze drying maintained a uniform porous network within the scaffold with pores ranging from 100–220 μm in diameter. These pores were much larger than the natural porosity of native collagen tissue but allowed for even nutrient diffusion and rapid cellular proliferation throughout the scaffold.
For applications when conventional collagen hydrogels are too weak, plastic compression of the collagen post-polymerization has been investigated (see Section 6.3.1). Through compression, water is expelled from the hydrogel to increase the concentration and density of collagen while also maintaining its fibril structure [647]. Although the general method for compressing collagen gels is similar across research groups (Fig. 11), the compression process has not been standardized; each group uses different load weights, load times, and compression apparatuses to achieve their desired end product. Many groups have reported quick processing times using custom-built load cells with compressive forces ranging from 1 [648] to 12.5 kPa [649]. Other groups used commercially available weights and culture dishes and therefore used longer compression times to compensate for lower compressive forces [650]. Furthermore, plastic compression of the collagen hydrogel can occur under the weight of the hydrogel itself [651]. Although natural compression eliminates the need for specialized equipment, this process occurs on the time scale of several hours, whereas external compression occurs over several minutes. Through plastic compression, collagen concentrations can be increased significantly and can reach concentrations over 10 [651] to 20 times [652] higher than the initial concentration of the gel. Furthermore, encapsulated cells remain viable throughout the process.
Fig. 11.
Generalized schematic of collagen gel compression apparatus
5.3. Temporal and spatial dynamic control of hydrogel properties
Compared with static hydrogel networks, dynamic control of hydrogel properties can provide desired release rates of cargo loaded in the hydrogel, regulate interactions with embedded cells, and create structural complexity within a scaffold to guide tissue regeneration [653–655]. The ECM is a dynamic environment with cells responding to changes in the ECM during processes such as wound healing, tissue development, and cancer progression [656]. Furthermore, the structure of the ECM is far more complex than synthetic hydrogel networks in most aspects, including physical, chemical, and biological cues [657]. To mimic native ECM properties and study dynamic biological processes, there is high demand for collagen and/or HA hydrogels with temporal and spatial properties that can be tuned dynamically. Stimuli-responsive chemistry and biofabrication technologies have emerged as powerful methods to dynamically manipulate hydrogel degradation, mechanical properties, and bioactivity, as well as enable on-demand changes in the microenvironment of gel matrices [658,659].
Many studies focus on tuning degradation rates of hydrogel networks [660–662]. Section 2 discusses the natural mechanisms for degrading collagen and HA and for clearing the degradation products. In particular, the body rapidly degrades unmodified HA [24]. It is preferable to control the degradation rate of collagen and HA in specific applications, such as drug delivery with prolonged release profiles or for aligning with tissue regeneration rates of the body [7,24,653,663,664]. To slow the degradation of collagen and/or HA hydrogels, various types of crosslinking methods have been employed. For example, glutaraldehyde crosslinking has been shown to resist enzyme degradation in collagenous materials [7,665]. EDC crosslinking of collagen allows for controlled degradation rates via variation of the degree of crosslinking [666]. Poly(hydroxy ester) modified methacrylated HA gels are hydrolytically degradable, and degradation was tailored through the density of ester groups [667].
Stimuli, including pH and UV light, as well as biological factors such as MMPs, glutathione (GSH), and ROS, have been applied to cleave stimuli-sensitive chemical bonds or specific peptide sequences to change degradation dynamically [17,668,669]. For instance, MMP-cleavable sequences have been added as crosslinkers in HA gels for controlled degradation [670]. Doxorubicin (DOX) was conjugated to thiolated HA, and the polymer solution was crosslinked by oxidation of thiol groups in air [671]. The cumulative release rate of DOX was increased by introducing GSH, which promoted cleavage of the disulfide bonds in the hydrogel matrix. The use of light allows for precise spatiotemporal control. Gao et al. developed a HA hydrogel with photo-responsive degradation properties [672]. HA grafted with coumarin and methacrylate formed a hydrogel under UV irradiation at 365 nm, whereas the hydrogel structure was partially degraded through exposure to UV light at 254 nm as coumarin decomposed. This hydrogel system modulated cell migration behavior in a controlled manner through in situ temporal control of crosslinking and subsequent substrate stiffness. Thus, the degradation rate of hydrogels can be controlled by incorporation of different crosslinking methods or stimuli-responsive modifications to meet the specific requirements of different applications.
Stimuli-responsive dynamic chemistries, such as reversible or switchable covalent bonds, ionic bonds, and host-guest bonds, have been investigated for preparing reversible hydrogels to capture ECM dynamics [533,534]. Gillette and co-workers created a two-component hydrogel in which collagen provided structural support and the alginate crosslinking state was reversibly switched (Fig. 12A) [386,673]. Specifically, a CaCl2 solution was used to crosslink alginate, whereas the addition of sodium citrate, a chelator of Ca2+, led to the reversal of alginate crosslinking. Fibroblast spreading and migration were restricted in the collagen-alginate gel when the alginate was crosslinked but was restored when the crosslinks were removed. This design allows for reversible 3D microenvironments that are suitable for studying changes in cell and tissue behavior. In another example, a dynamic tunable HA hydrogel was synthesized using supramolecular crosslinks through azobenzene bound to β-cyclodextrin [459]. Upon irradiation at 365 nm, softer gels resulted because azobenzene isomerized to the cis configuration and had decreased interactions with β-cyclodextrin. The network connectivity was restored by irradiation with visible light (400–500 nm), which reverted azobenzene to the trans state (Fig. 12B). The storage modulus of the hydrogel changed rapidly in the presence of different wavelengths, and multiple cycles indicated repeatable reversibility.
Fig. 12.
Reversible modulation of hydrogel properties using stimuli-responsive bonds. (A) dynamic crosslinking state of alginate in collagen-alginate composites via treatment with Ca2+ or sodium citrate. Reprinted with permission from Gillette et al., 2010 [386]. Copyright 2010 John Wiley and Sons. (B) Photoreversible modulation of matrix mechanics based on the host-guest pairing of azobenzene and β-cyclodextrin-containing HA. Reprinted with permission from A.M. Rosales, C.B. Rodell, M.H. Chen, M.G. Morrow, K.S. Anseth, J.A. Burdick, Bioconjugate Chem. 29 (2018) 905–913. Copyright 2018 American Chemical Society [459].
As introduced in section 5.2, many processing techniques have been utilized to prepare hydrogels with desired structures. Hadjipanayi reported a collagen matrix with a continuous directional stiffness gradient fabricated by using a horizontal plate to compress collagen gels into a wedge shape [16]. The collagen density gradient after compression corresponded to the stiffness gradient. Substrate stiffness profoundly affects cell behavior, and cells seeded in this matrix tended to accumulate towards the stiff region of the gradient.
Multiple modes of crosslinking applied sequentially and in combination with mask-based photolithography have been used to spatially modulate the properties of hydrogels [674–676]. Specific control of photoinitiator concentration, light intensity, and irradiation time is required to maintain pattern fidelity and improve the cell compatibility of the gels. To investigate the effect of local mechanics on MSC behavior, methacrylated HA was crosslinked sequentially via Michael-type addition and UV-initiated radical polymerization [677]. By restricting the UV exposure regions and varying UV irradiation time, a wide range of stiffnesses was prepared within individual gels, and the MSCs exhibited spreading and proliferation behavior corresponding to the local stiffness gradient of each gel.
Recently, 3D bioprinting technologies have provided effective alternatives to photopatterned hydrogels with high complexity and precision in a volumetric space (see section 5.2.2) [659]. Mohamed and colleagues provide an in-depth review of preparing in vitro dynamic hydrogels via photolithography and 3DBP [678]. Integration of stimuli-responsive materials with 3DBP results in hydrogel scaffolds capable of time-dependent transformation and is commonly defined as 4D bioprinting.
Strategies to engineer collagen and/or HA hydrogels through a combination of chemical modifications and technological platforms have attracted much interest. Advancing the ability to fabricate dynamic hydrogels will enhance our capability to explore and guide cellular behavior, tissue regeneration, and disease therapy.
6. Biomedical applications of collagen and HA hydrogels
Collagen and/or HA hydrogels are used extensively in the medical field for applications such as tissue engineering and developing in vitro tissue models because both collagen and HA are important components of numerous tissues as described by this review. Researchers vary aspects of the hydrogels, such as collagen type and presentation or HA crosslinking method, to tune the properties to match specific mechanical properties and functions of the tissue. In Section 3.3, we discussed current commercial applications, whereas, in this section, we discuss a few notable examples of recent developments for collagen and HA hydrogels for bone, cartilage, and skin tissue engineering and for in vitro disease models.
6.1. Bone tissue engineering
The healing of bone is impaired in the case of severe injuries, osteogenesis imperfecta, congenital malformation, osteoporosis, or rheumatoid arthritis, and external intervention can initiate and escalate bone repair and regeneration. Autografts, allografts, and xenografts are the traditional methods used for healing bone defects. However, their clinical use is limited due to donor site morbidity, foreign body reaction, and low availability of tissue. Hydrogels used for bone tissue engineering create a favorable environment for drug delivery and to heal bone defects by encapsulating cells to promote cell attachment, migration, proliferation, and differentiation (Fig. 13) [356,679].
Fig. 13:
Schematic representation of bone tissue engineering using nanostructured scaffolds. [680]. Reprinted from Biomaterials 31, L. Wang and J.P. Stegemann, thermogelling chitosan and collagen composite hydrogels initiated with β-glycerophosphate for bone tissue engineering, 3976–3985, copyright (2010), with permission from Elsevier.
6.1.1. Collagen hydrogels
Collagen I is biocompatible, has osteoinductive properties, and forms scaffolds that are overwhelmingly used for bone regeneration [681–683]. Many of these scaffolds are discussed in section 3.3.1.2. However, improvements are still needed due to the lack of mechanical strength and rapid degradation of collagen fibers by collagenase enzymes. One approach has been to incorporate additional polymeric materials to increase the strength and stability of collagen hydrogels. For example, Gurumurthy et al. demonstrated that a composite hydrogel of collagen-ELP containing Bioglass particles supported in vitro osteogenic differentiation of stem cells and in vivo healing of critical-sized cranial bone defects in a rat model [684].
Collagen-based hydrogels have also been explored for developing injectable hydrogels. One example is a collagen/chitosan-based injectable hydrogel, where β-glycerophosphate was incorporated to catalyze gelation of the composite at physiological pH and temperature [680]. Human bone marrow-derived stem cells were easily incorporated in these hydrogels at the time of gelation and expressed increased levels of osteogenic markers. Huang et al. also developed injectable hydrogels composed of collagen, nanohydroxyapatite, and chitosan for bone defect repair, and the microstructure was similar to physiological bone [685].
6.1.2. HA hydrogels
HA-based hydrogels are widely employed in bone tissue engineering due to their biocompatibility and ability to support cellular proliferation and migration. In this context, Zhou et al. reported a photo-crosslinked bioactive hydrogel composed of methacrylated HA and an arginine-based unsaturated poly(ester amide) [686]. The compressive modulus and swelling capacity of the hydrogel could be modulated as a function of polymer content. Enhanced expression of bone-specific genes and promising bone regeneration demonstrated the efficacy of the hydrogel for bone tissue engineering.
HA-based hydrogels not only have been used to accelerate cellular proliferation and necessary gene expression but also have supported the sustained release of drugs. For example, a thiol-modified HA-based hydrogel Glycosil™ (BioTime Inc., USA) manifested an initial burst release followed by a more sustained release of BMP-2 and thus prevented the adverse effects of BMP-2 overdose [687]. In a study by Patterson and colleagues, BMP-2 and vascular endothelial growth factor (VEGF) were co-delivered in a sustained manner to rat cranial bone defect sites via glycidyl methacrylate-modified HA hydrogels and thus resulted in significant bone mineralization and repair [660]. In a similar approach, Holloway et al. developed maleimide-modified HA crosslinked with MMP-sensitive cell adhesion peptides to allow protease-mediated hydrogel degradation and BMP-2 delivery [688]. Their results suggested the rate of hydrogel degradation and subsequent growth factor release had a significant influence on mineralization and bone formation in rat cranial defects. In a peri-implant application, zoledronate, a bisphosphonate medication used for treating bone disease, and hydroxyapatite nanoparticles were co-delivered via a HA hydrogel to augment bone while inhibiting bone resorption during osteoporosis [689].
6.1.3. ColHA hydrogels
Attempts to combine collagen and HA have demonstrated better effects than using either one alone. For example, Liu and colleagues crosslinked activated formyl group-modified HA with collagen to form a new ColHA matrix that resulted in improved bone healing and osteoconduction in the rat skull bone as compared to collagen or hyaluronate alone [690]. Silica-based materials play a significant role in bone formation by stimulating collagen synthesis and matrix biomineralization [691]. Thus, surface-modified silica particles were dispersed in a collagen-chitosan-HA solution and crosslinked via genipin to result in a biocompatible, bioactive, injectable hydrogel suitable for osteoblast mineralization [568]. A similar approach was previously reported by Gilarska et al. for developing a genipin-crosslinked injectable hydrogel from collagen-chitosan-HA, and the hydrogel supported cellular proliferation and adhesion [567]. Zhang and coworkers fabricated a photochemically crosslinked hydrogel containing glycidyl methacrylate-modified collagen and methacrylic anhydride-modified HA [556]. The hydrogel promoted osteogenic differentiation of encapsulated stem cells without the usage of BMP-2.
Bone tissue engineering is a rapidly growing field with newly developed materials boasting improved and faster bone repair and regeneration. Many reviews provide excellent insights into the evolution and methodologies adopted to design collagen and HA-based hydrogels for bone tissue repair [356,679,692,693]. Overall, ColHA hydrogels, with their additional functionalities and the ability to serve as a matrix for encapsulating cells and drugs, can be beneficial for bone regeneration.
6.2. Cartilage tissue engineering
Articular cartilage has limited ability for self-repair since it is an avascular and nearly acellular tissue [24,36,359]. Thus, because it is difficult for repair cells to access cartilage, tissue engineering is an attractive option for stimulating cartilage regeneration in clinical diagnoses such as acute, traumatic sports injuries and osteoarthritis.
Cartilage is primarily composed of water, comprising 80% of the tissue wet weight [36]. Of the dry components, collagen II accounts for ~90–95% of the collagen present in cartilage ECM. Other major components of cartilage include proteoglycans, GAGs, smaller matrix proteins, lipids, and chondrocytes. Given their abundance in cartilage ECM, collagen and HA are both attractive options for use in cartilage tissue engineering [694].
6.2.1. Collagen hydrogels
Collagen I was among the earliest natural polymers to be used for cartilage tissue engineering since it is relatively inexpensive and self-assembles into hydrogels at physiological conditions without the addition of chemical crosslinks. Collagen I hydrogels have been studied since 1994 with various combinations of cell types and animal or human models [695–699]. As described in Section 3.3.1.2, collagen I continues to be the primary material used in MACI procedures. Some drawbacks to using collagen I alone for cartilage tissue engineering are that it can elicit undesirable phenotypes, such as osteogenic, in stem cells and cause dedifferentiation of chondrocytes [694]. Additionally, collagen hydrogels are often softer (Young’s modulus ~65 kPa) than native cartilage [663,694].
Collagen II is found natively in cartilage and therefore confers desirable biological cues to cells encapsulated in hydrogels. However, it is often not utilized alone due to its higher cost and less robust hydrogel formation [535,551,700]. To remedy these limitations, collagen I can be employed as the primary scaffold component to create robust hydrogels and lower costs, and collagen II can be added in smaller quantities. For example, a recent study used a blend of collagen I and II and found that, compared to hydrogels composed only of collagen I, the blended hydrogel promoted increased in vitro production of cartilage matrix and superior in vivo cartilage regeneration [701]. However, there are a few examples of hydrogels composed only of collagen II, and these hydrogels showed better chondrogenic stimulation than collagen I hydrogels [702].
As mentioned in Section 3.3.1.2, collagen I and III are used clinically in different countries in autologous chondrocyte implantation applications such as MACI, Maix, and Chondro-Gide [359]. The choice of collagen I and III over collagen II, the primary collagen type found in cartilage, was made to support the idea of introducing cells in a less-differentiated, more motile state [703]. The authors note that although collagen II promotes stable chondrocyte phenotypes, the cells may have a lower capacity for division, which is undesirable when trying to integrate matrix into host cartilage.
6.2.2. HA hydrogels
As detailed in Section 4.3, crosslinked HA is used to lubricate joints and alleviate pain for those suffering from osteoarthritis, and HA has been extensively studied for use in cartilage tissue engineering due to its inherent biological and mechanical cues that are desirable for cartilage generation. These HA cues have been shown to lead to chondrogenic differentiation of stem cells to support cartilage regeneration [704,705]. In an attempt to further enhance cartilage tissue engineering, numerous crosslinking modifications have been used for HA hydrogels, such as methacrylation [561,704,706,707], hydrazone covalent bonding [707,708], and other covalent crosslinking methods [706,709,710], to modulate the mechanical properties of the hydrogels to match those of native cartilage.
6.2.3. Blended hydrogels
Various blended hydrogels, or multi-component hydrogels, have been studied for cartilage tissue engineering applications. Multiple types of collagen have been blended together with seeded chondrocytes [711] or stem cells [535,701]. Collagen has been combined with GAGs, such as HA and/or CS [561,562,712,713], or synthetic polymers [541,714]. CS is a common choice for blended hydrogels for cartilage applications because it participates in biological processes such as providing anti-inflammatory effects in osteoarthritis, reducing the friction coefficient of articular cartilage, and promoting the secretion of proteoglycans by chondrocytes [715,716]. Numerous groups investigated HA hydrogels with different crosslinking materials, such as PEG [717–719] and polyvinyl alcohol [720], or the impact of different peptide groups, such as the RGD cell-binding domain or MMP-sensitive sequences [670,705,708]. The motivation behind the use of blended hydrogels is to incorporate native components of cartilage that provide biological cues to seeded cells and modulate crosslinking densities to more accurately recapitulate the native cartilage environment and its mechanical properties. Additionally, when comparing blended hydrogels to single component hydrogels, the blended gels often result in better chondrogenic differentiation [535,701,713].
Cartilage tissue engineering has been studied for over thirty years; however, new findings about the importance of factors such as dynamic loading and the implications of inflammation continue to advance the field [721,722]. The state of cartilage tissue engineering has been reviewed extensively within the last decade [359,721,723,724]. Natural polymers used for cartilage tissue engineering applications have been reviewed as well [663,694,725–727].
6.3. Skin tissue engineering
Full thickness skin defects have difficulty healing naturally due to their size and loss of vasculature, and these wounds are also highly susceptible to bacterial infections. Such wounds can be found in patients suffering from extensive third-degree burns, physical trauma, or surgical procedures, including tumor resections. The natural healing process can also be complicated by conditions such as diabetes mellitus in which wound healing pathways are further hindered due to abnormalities in the vascular system [728]. While skin autografts and allografts can be used to protect the wounds and promote healing, these materials are in short supply [729]. As such, many skin tissue engineering approaches have been developed to address the need for skin regeneration and commonly use collagen and HA due to their natural occurrence in skin tissue as well as their bioactive properties. Section 3.3.1.1 discussed some current commercial products, whereas this section discusses new advances.
6.3.1. Collagen hydrogels
Due to the poor mechanical properties of collagen-only hydrogels [730], many skin tissue engineering approaches incorporate some method to strengthen the collagen and create a substrate that can be easily handled and maintain its size without significant contraction. By resisting contraction, reinforced collagen hydrogels are able to maintain coverage of the wound site throughout the duration of the healing process.
Collagen-based composite materials have been explored to strengthen hydrogels. Using commercially available meshes, Hartmann-Fritsch et al. were able to strengthen collagen hydrogels through casting the gels around either trimethylene carbonate and lactide copolymer meshes or poly(lactic-co-glycolic acid) meshes, both of which allowed the constructs to be easily handled by human operators while also being completely biodegradable [731]. Another approach applies bioconjugate chemistry to covalently crosslink collagen fibrils during gelation and thus strengthen the hydrogel while preserving some of the natural fibril structure. Using four-armed PEG succinmidyl glutarate as a crosslinker, Lotz et al demonstrated a biocompatible in situ gelling hydrogel that was able to resist contraction via encapsulated fibroblasts over the course of twenty days, whereas un-crosslinked gels contracted to half their size [732]. Furthermore, compared to the unmodified gel, the increase in stiffness due to the crosslinks also promoted keratinocyte proliferation, which was demonstrated by an increase in Ki67 positive cells.
Plastic compression has emerged as a method for quickly processing cell-laden collagen hydrogels for skin tissue engineering purposes. By increasing the collagen fibril density through compressive force, a stronger scaffold can be fabricated quickly, with procedures taking between 5 and 15 minutes [649,733–737]. Conversely, collagen hydrogels contracted through cell culture require several days of cell encapsulation to reach similar densities. Plastic compression has allowed for the rapid creation of collagen hydrogels with stiffnesses and tensile strengths close to skin while also maintaining high cell viability since the majority of encapsulated cells survive the compression process.
Using this process, Braziulus et al. achieved collagen hydrogels with concentrations of collagen as high as 65 mg/mL [734]. Similarly, Sohutskay et al. reported post-compression concentrations of collagen of 20 mg/mL and 40 mg/mL [737]. In all of these cases, increases in collagen concentration were accompanied by increases in substrate stiffness and ease of handling. Compared to the uncompressed, low-density collagen hydrogels, compressed collagen hydrogels seeded with fibroblasts and keratinocytes showed better skin healing outcomes, such as host integration and revascularization of the wound [737]. Such effects are attributed to the increased stiffness of compressed collagen hydrogels, compared to uncompressed gels and commercially available collagen sponges, since the response of dermal fibroblasts and keratinocytes is more favorable when substrate stiffness matches more closely the stiffness of the native tissue (~1 MPa) [649].
6.3.2. HA hydrogels
HA-based materials have also been investigated as possible cell scaffolds for skin wound healing as HA retains large volumes of water and has been shown to accelerate the wound healing process [738]. However, due to the lack of available cell-binding sites and its susceptibility to degradation, most HA hydrogels for skin tissue repair have been composite materials. Secondary components that have been studied include gelatin [739], collagen [555], and decellularized extracellular matrices [740], and hydrogels containing a combination of these components and HA all decrease wound healing time and increase angiogenesis at the wound site. Other polymers included in HA hydrogel composites include alginate [741], for increased stiffness, and chitosan [742], as an antimicrobial additive.
In addition, HA-based materials have often been used as delivery vehicles for other agents to promote wound healing. Examples include growth factors such as VEGF, basic fibroblast growth factor [743,744], and keratinocyte growth factor [745]. Similarly, antibiotics such as vancomycin have been loaded into these gels to prevent bacterial infection while the wound is still healing [746]. Due to the negative charge and high hydrophilicity of HA, these agents can be slowly released into the surrounding environment and provide sustained local concentrations for the promotion of angiogenesis and wound closure.
Although many commercially available collagen and HA products are available for skin tissue repair, as detailed in Section 3.3, the field of skin tissue engineering continues to evolve. Through modulating stiffness and microstructure or including other bioactive agents, collagen and HA hydrogels can be designed for faster wound closure rates and improved quality of engineered skin tissue.
6.4. Engineered cancer models
Recapitulating the tumor microenvironment (TME) is a common area of investigation for scientists and engineers who are developing innovative and clinically-relevant models for the prediction of clinical outcomes and therapeutic efficacy. The TME includes but is not limited to: the extracellular matrix, unique populations of normal and cancer cells, genetic factors, and sequestered cytokines that have a direct impact on tumor behavior [747]. Tumor heterogeneity, a fundamental characteristic of the TME, describes the variation in these factors that makes cancers unique within and between different patients [748,749]. Despite the recent developments in the field, our understanding of how ECM tumor heterogeneity affects cancer progression, drug resistance, and therapeutic response remains limited.
ECM components such as collagen and HA are primary targets of study for understanding how heterogeneity drives disease progression. Collagen is relevant structurally and biochemically for cell invasion and metastasis. HA influences molecular transport of cytokines and growth factors via tissue hydration and viscoelasticity throughout the TME and alters tissue microstructure, mechanics, and perfusion [201,578,750]. Furthermore, the known interactions of HA with CD44 and RHAMM receptors are thought to lead to invasion and metastasis in some cancers [751,752]. While these phenomena are well reviewed, the primary focus of the following sections will be on studies that focus on interactions between collagen, HA, and other ECM components. Literature reviews discussing collagen [753,754] and HA [755–757] in cancer may provide a more in-depth commentary on the individual contributions of each matrix component towards disease progression.
6.4.1. ColHA in vitro models for breast cancer heterogeneity
The creation of physiologically relevant models is important for investigating the TME. As vital ECM components, collagen and HA both play a role in the study and treatment of breast cancer, the most common cancer among women and a contributor via metastasis toward lung cancer, the leading cause of cancer-related mortality [758–760]. Stromal HA is a potential predictor for low overall survival and lymph node metastasis, and its presence has been shown to be upregulated in tumorous tissues [761,762].
Many models within this research area utilize chemical crosslinking methods to enable tunability. Bonnesoeur and coworkers prepared HA hydrogels pre-functionalized with poly(L-lysine), collagen III, or collagen IV as cellular recognition domains [763]. They observed an increase in viability for MDA-MB-231 breast cancer cells with increasing genipin concentrations, and these results indicate that crosslink density and pore diameter influence cellular behavior. Methacrylated HA has been used for studying the effects of dynamic stiffening and hypoxia on the epithelial to mesenchymal transition, the process where cells lose their polarity and gain enhanced expression of new transcription factors and a migratory phenotype [764,765]. Similarly, cellular invasion was investigated using a Diels-Alder crosslinked HA functionalized with protease sensitive peptides, and these studies showed, once again, that motility depends on matrix density and cellular traction in these hydrogel systems [661]. These models lend significant insight toward the investigation of cancer cell lines in vitro; however, they remain mostly specific to the conditions of the study being conducted.
To develop a more generalized mammary gland model, Campbell and coworkers utilized freeze drying and EDC crosslinking of collagen in the presence of 7.5% or 15% HA for the creation of a tissue model with epithelial and stromal cells [766]. Whereas the model by Campbell does not specifically address metastatic disease, it may optimally serve as the basis for many breast tumor models since the platform maintains stable morphology with crosslinked HA over extended durations, is amenable to biochemical and mechanical manipulation, and enables inclusion of primary human cells for personalized medicine.
Although formulated hydrogels are useful as platforms for investigation, in vitro platforms utilizing endogenously produced ECM offer increased fidelity for understanding physiological processes. A model created by Mazio et al. demonstrated the heterogeneity between tumors and featured matrix conditions that changed over time by including cells, such as MCF-7 breast adenocarcinoma cells, human umbilical vein ECs, and human dermal fibroblasts, that produce ECM [767]. Direct synthesis of endogenous collagen and HA by cells allowed for the identification of the driving factors of matrix production and the downstream effects within the tumor microenvironment. Higher collagen deposition, fiber alignment, and HA production was found in the ECM produced by cancerous cells, as compared to non-cancerous controls, and these results indicate the importance of these features in contributing to the desmoplastic morphological changes of the TME in human breast cancer [767].
6.4.2. Collagen/HA models with the inclusion of alternative ECM components
Although collagen- and HA-based in vitro models are crucial platforms for studying the effects of the ECM on tumor progression, these platforms fail to address other aspects of the heterogeneous tumor microenvironment. It is important to include other ECM and related microenvironmental structures when studying ECM effects on an ever-changing disease that is possibly unique within one patient and between multiple patients. Similar to the tissue-altering properties of HA and collagenous tissue, fibrous fibronectin has been shown to be a key contributor to tissue stiffening and a contributor to the dissemination and proliferation of cells from primary tumors to secondary sites in breast cancer [765,768]. Shinde and coworkers showed that fibronectin ECM deposited by mesenchymal cells aids in the metastatic process for metastasis-competent cells within the microenvironment [769]. Furthermore, HA alters fibrous ECM production of fibrous fibronectin and fibrous collagen in lung myofibroblasts [770].
In breast cancer, tissue stiffening is associated with increased ECM production and poor patient survival [771]. Indeed, Gioiella and colleagues presented a microfluidic breast tumor platform that highlights the overproduction of HA and fibronectin in a reactive cancer microenvironment containing normal and cancer-activated fibroblasts with MCF7 malignant breast cancer cells [772]. The authors indicated that the study of these molecules as assembled ECM structures is vital to understanding tissue properties related to invasion and malignancy, yet few hydrogel models exist specifically investigating collagen, HA, and fibronectin.
It is known that the interplay between different ECM proteins, GAGs, and other ECM molecules can drive unique cellular phenotypes and tissue properties. When investigating the intricacies of cancer, there are limitless interactions and effects to study. Producing models containing collagen and HA along with other relevant ECM proteins can potentially enable a new understanding of heterogeneity in the breast tumor microenvironment and aid in the production of useful therapeutic discoveries and diagnostic platforms.
6.5. Other applications
Due to the ubiquity of collagen and HA molecules in ECM across various tissues, ColHA scaffolds have been applied to many other fields besides those discussed in this review. One prominent example is neural tissue engineering, including topics such as enhanced differentiation of neural stem/progenitor cells [773], peripheral nerve regeneration [612], repair of neural damage [774], and neural disease models for glioblastoma [775]. ColHA hydrogels have also been used to support cell culture, including but not limited to the differentiation of induced pluripotent stem cells to neuronal production [776], preadipocyte culture for adipose tissue engineering [646], and adipose-derived MSC culture for vocal fold regeneration [777]. Other applications of these hydrogels include use in corneal defect fillers [778], nucleus pulposus regeneration [536,779], DNA delivery systems [780], and immunoprotected islet transplantation [781]. The vast applications of collagen and hyaluronan matrices demonstrate their importance to the field of tissue engineering and regenerative medicine.
7. Conclusion and future directions
Collagen and hyaluronic acid represent important components of the extracellular matrix due to their structural and biochemical significance for tissue integrity and numerous cellular processes. As such, these materials have extraordinary relevance toward recapitulating physiological environments for biomedical applications. Work has been done to source these biopolymers from animal, human, bacterial, and recombinant methods; however, significant variation exists across different sources. This variation has a severe impact on the function in hydrogels because of differences in collagen structure, collagen post-translational modifications, and HA molecular weight. One solution to this biological variation exists in the development of a wide set of parameters that control hydrogel properties. Concentration, pH, chemical crosslinkers, and chemical modifications have been explored to control collagen microstructure features, such as fiber diameter and length, pore density, and size, as well as hydrogel properties including swelling capacity, biocompatibility, and degradation. Furthermore, hydrogels with chemically modified collagen and HA represent a deep repository of unique hydrogel compositions that can be flexibly tuned toward specific applications and biomedical questions. In seeking answers, researchers have extensively employed collagen and HA in hydrogels for bone, cartilage, and neural tissue cellular differentiation, full thickness skin defect repair, and disease models of cancer microenvironments.
Collagen and HA are ubiquitously present throughout biology, and therefore investigators have a wide range of desired goals and outcomes when creating ColHA hydrogels for biomedical research. Despite significant progress in engineering these hydrogels for specific applications, the immense flexibility of these materials provides a challenge in standardization and comparison of results given that blend ColHA hydrogels can exist with any number of unique component ratios, modifications, and crosslinkers. In addition, existing literature does not document the mechanisms by which collagen and HA interact within all unique tissues throughout the body to create specific structural and biochemical microenvironments. To address this challenge, establishment of standard properties for each tissue type with respect to collagen and hyaluronic acid would prove advantageous for choosing appropriate fabrication parameters in creating hydrogel mimics. Tools such as histology and mass spectrometry have the potential to describe the variation found in different tissue spaces. Armed with this knowledge, researchers can better develop strategies, such as particular crosslinking methods or processing techniques, that result in a suitably representative hydrogel system for the tissue or process of interest. Finally, there exist subtle differences between hyaluronic acid and other relevant GAGs, such as chondroitin sulfate and heparin sulfate, that also play an important role in collagen-containing hydrogels. Investigation of engineered hydrogels systems including these molecules could provide relevant interactions with collagen affecting assembly, structure, and function.
Acknowledgments
The authors would like to thank Dr. Michael Ladisch for his comments and suggestions for the manuscript. This work was supported by Eli Lilly and Company. This work was jointly supported by an NIH training grant (T32 HL086350 to support MN), an NSF graduate research fellowship (for JT), and a Purdue Frederick N Andrews Fellowship (for PMB).
Abbreviations
- ECM
extracellular matrix
- HA
hyaluronic acid
- GAG
glycosaminoglycan
- ColHA hydrogel
collagen and HA blend hydrogel
- COL
collagenous
- NC
non-collagenous
- FACIT
fibril associated collagens with interrupted triple helices
- O
hydroxyproline
- CMP
collagen mimetic peptide
- Tm
melting temperature
- MMP
matrix metalloproteinase
- MW
molecular weight
- GlcNAc
N-acetyl-D-glucosamine
- HAS
hyaluronan synthase
- ROS
reactive oxygen species
- DDR
dimeric discoidin receptor
- GPVI
glycoprotein VI
- LAIR-1
leukocyte-associated immunoglobulin-like receptor-1
- ECs
endothelial cells
- RHAMM
receptor for HA-mediated mobility
- HARE
hyaluronan receptor for endocytosis
- CS
chondroitin sulfate
- GRAS
generally recognized as safe
- (rh)BMP-2
(recombinant human) bone morphogenetic protein 2
- FDA
Food and Drug Administration
- MACI
matrix-assisted autologous chondrocyte implantation
- MSCs
mesenchymal stem cells
- PEG
poly(ethylene glycol)
- EDC
1-ethyl-3-(3- dimethylaminopropyl)carbodiimide
- GA
glutaraldehyde
- NHS
N-hydroxysuccinimide
- GP
genipin
- CA
citric acid
- ELP
elastin-like protein
- BDDE
1,4-butanediol diglycidyl ether
- DA
Diels-Alder
- HRP
horseradish peroxidase
- IEDDA
inverse electron-demand DA
- PDMS
poly(dimethylsiloxane)
- 3DBP
three dimensional bioprinting
- UV
ultra-violet
- GSH
glutathione
- DOX
doxorubicin
- VEGF
vascular endothelial growth factor
- TME
tumor microenvironment
Author biography

Dr. Qinghua Xu received her PhD degree in Polymer Chemistry from Changchun Institute of Applied Chemistry, University of Chinese Academy of Sciences in 2017. She then worked as a postdoctoral researcher from 2017–2020 at Cornell University. She is currently a postdoctoral researcher under the supervision of Dr. Julie C. Liu at Purdue University. Her research interests are synthetic and natural polymer-based nanoparticles and hydrogels for drug delivery and tissue engineering. Her current research focuses on in vitro adipose tissue modeling.

Jessica Torres is a PhD student in the Davidson School of Chemical Engineering at Purdue University. She received her B.S. in chemical biological engineering from the Massachusetts Institute of Technology (MIT) in 2016. She received a 2019 National Science Foundation (NSF) Graduate Research Fellowship to support her studies. Her research focuses on the development of protein-based biomaterials for biomedical applications including tissue sealants and adhesives and in vitro tissue models for drug delivery.

Mazin Hakim is a PhD student in Biomedical Engineering under the supervision of Professor Luis Solorio. He received his B.A. in Chemistry with a minor in Mathematics from Wabash College in 2017, where he participated in research involving 3D printing of low cost chemical laboratory instruments. His current research aim is focused on understanding and optimizing subcutaneous drug delivery of biotherapeutics.

Paulina M Babiak is a Ph.D. student in chemical engineering under the supervision of Professor Julie C. Liu at Purdue University. She is a 2021 NSF Graduate Research Fellowship recipient. She received her B.S. in chemical engineering from Columbia University in 2019, with minors in biomedical engineering and East Asian studies. Her research focuses on in vitro adipose tissue modeling and recombinant protein materials.

Dr. Pallabi Pal received her Ph.D. in Biomaterial Science and Tissue Engineering from Indian Institute of Technology Kharagpur, India in 2017. She then worked as postdoctoral researcher from 2017–2020 at the University of Mississippi Medical Center. She is currently working in Purdue University as postdoctoral researcher. Her research focuses on effect of hyaluronic acid molecular weight and concentration on chondrogenesis.

Carly Battistoni is a Ph.D. student in Chemical Engineering under the supervision of Dr. Julie C. Liu at Purdue University. She received her B.S. in Chemical and Biomolecular Engineering from the University of Delaware, with a minor in Biochemical Engineering. Her current research focuses on cartilage tissue engineering.

Michael Nguyen is a Ph.D. candidate in the biomedical engineering graduate group at the University of California, Davis. He obtained his B.S. in biomedical engineering at Arizona State University in 2018. He is currently a member of the Laboratory for Engineered Therapeutics and under the supervision of Dr. Alyssa Panitch. His research focuses are glycosaminoglycan-based biomaterials and proteoglycan mimetics for tissue engineering and regenerative medicine purposes.

Dr. Alyssa Panitch received bachelor’s degrees from Smith College in Biochemistry and from the University of Massachusetts-Amherst in Chemical Engineering. She completed her Ph.D. in Polymer Science and Engineering from the University of Massachusetts. After a postdoctoral fellowship at the Swiss Federal Institute of Technology (ETH) and University of Zurich, she accepted a position of Assistant Professor in the Harrington Department of Bioengineering at Arizona State University. In 2006 she joined the faculty in the Weldon School of Biomedical Engineering at Purdue University and in 2013 was named the Leslie A. Geddes Professor of Biomedical Engineering. In January 2015 she accepted a position as Vice Provost for Faculty Affairs at Purdue University. In this position, she oversaw faculty hiring and retention for the university, updated the promotion and tenure policies, procedures and guidelines, and led professional development efforts. In June 2016, she was appointed as the Edward Teller Professor and Chair of Biomedical Engineering at University of California Davis, and she currently serves as the Executive Associate Dean and the Associate Dean for Academic Personnel and Planning in the College of Engineering at the University of California Davis. In her current position, she oversees the merit and promotion process and faculty and academic personnel hiring for the college. She is a member and Fellow of the Biomedical Engineering Society, the American Institute for Medical and Biological Engineers (AIMBE), and the National Academy of Inventors. She also serves as the Secretary and Treasurer and on the Executive Board for AIMBE and as an Editor for the Journal of Colloids and Surfaces B: Biointerfaces.

Dr. Luis Solorio, Ph.D., is the director of the Tumor Microenvironment & Therapeutics Lab (TMET) at Purdue University, which focuses on applying principles of tissue engineering, medical imaging, and drug delivery for the development of modular 3D tissue-engineered constructs that can be used to evaluate the cancer cell response to microenvironmental cues. He joined the faculty at Purdue University in 2016 as an Assistant Professor in the Weldon School of Biomedical Engineering. He is a U.S. Army veteran who proudly served for 5 years immediately after graduating high school. Dr. Solorio has been trained in both engineering and chemistry, obtaining a B.S. in Biomedical Engineering and Chemistry from Saint Louis University in St. Louis in 2006. Dr. Solorio received his M.S. degree in Biomedical Engineering from Rensselaer Polytechnic Institute in 2007 working with Dr. Jan Stegemann exploring methods of growth factor delivery to drive differentiation of mesenchymal stem cells. Dr. Solorio then received his Ph.D. in Biomedical Engineering from Case Western Reserve University in 2012 with Dr. Agata Exner focusing on the use of medical imaging to guide the design and development of controlled release platforms. He then did his postdoctoral research at the University of Michigan under the mentorship of Dr. Joerg Lahann and Dr. Gary Luker where he developed novel platforms for studying the tumor microenvironment and was the recipient of the K99 Pathway to Independence Award.

Dr. Julie C. Liu received her B.S.E. in chemical engineering from Princeton University. She was awarded a Whitaker Graduate Fellowship for her doctoral research with Dr. David Tirrell at the California Institute of Technology. She received an NIH postdoctoral fellowship for her research at the University of Massachusetts Medical School with Dr. Jane Lian and Dr. Gary Stein. She is an associate professor of chemical engineering and biomedical engineering at Purdue University. She is currently a Purdue University Faculty Scholar and won the 2021 Purdue College of Engineering Faculty Excellence Award for Graduate Student Mentorship. She was selected as a 2019–2021 fellow in the Executive Leadership in Academic Technology, Engineering, and Sciences (ELATES) program. Her research interests include biomimetic materials, tissue engineering, stem cell differentiation, and surgical adhesives. Her research has been funded by the National Science Foundation, National Institutes of Health, Department of Defense, American Heart Association, and a 3M Nontenured Faculty Award.
Footnotes
Declaration of Competing Interest
The authors declare no competing financial interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Bello AB, Kim D, Kim D, Park H, Lee S-H, Tissue Eng. Part B Rev. 26 (2020) 164–180. [DOI] [PubMed] [Google Scholar]
- [2].Kleinman HK, Philp D, Hoffman MP, Curr. Opin. Biotechnol. 14 (2003) 526–532. [DOI] [PubMed] [Google Scholar]
- [3].Xing H, Lee H, Luo L, Kyriakides TR, Biotechnol. Adv. 42 (2020) 107421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Theocharis AD, Skandalis SS, Gialeli C, Karamanos NK, Adv. Drug Deliv. Rev. 97 (2016) 4–27. [DOI] [PubMed] [Google Scholar]
- [5].Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J, in: Molecular Cell Biology, 4th ed., W. H. Freeman; New York, 2000. [Google Scholar]
- [6].Ricard-Blum S, Cold Spring Harb. Perspect. Biol. 3 (2011) a004978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Friess W, Eur. J. Pharm. Biopharm. 45 (1998) 113–136. [DOI] [PubMed] [Google Scholar]
- [8].Wang T, Lew J, Premkumar J, Poh CL, Win Naing M, Eng. Biol. 1 (2017) 18–23. [Google Scholar]
- [9].Luo T, Kiick KL, Bioconjugate Chem. 28 (2017) 816–827. [DOI] [PubMed] [Google Scholar]
- [10].Meyer M, Biomed. Eng. OnLine 18 (2019) 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Burdick JA, Prestwich GD, Adv. Mater. 23 (2011) H41–H56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Highley CB, Prestwich GD, Burdick JA, Curr. Opin. Biotechnol. 40 (2016) 35–40. [DOI] [PubMed] [Google Scholar]
- [13].Fratzl P, in: Fratzl P (Ed.), Collagen: Structure and Mechanics, Springer US, Boston, MA, 2008, pp. 1–13. [Google Scholar]
- [14].Tsai S-W, Liu R-L, Chen C-C, Biopolym. 83 (2006) 381–388. [DOI] [PubMed] [Google Scholar]
- [15].Yang Y, Kaufman LJ, Biophys. J. 96 (2009) 1566–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hadjipanayi E, Mudera V, Brown RA, Cell Motil. 66 (2009) 121–128. [DOI] [PubMed] [Google Scholar]
- [17].Suo A, Xu W, Wang Y, Sun T, Ji L, Qian J, Carbohydr. Polym. 211 (2019) 336–348. [DOI] [PubMed] [Google Scholar]
- [18].Sato N, Taniguchi T, Goda Y, Kosaka H, Higashino K, Sakai T, Katoh S, Yasui N, Sairyo K, Taniguchi H, J. Proteome Res. 15 (2016) 4709–4721. [DOI] [PubMed] [Google Scholar]
- [19].Frantz C, Stewart KM, Weaver VM, J. Cell Sci. 123 (2010) 4195–4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Myers RB, Fredenburgh JL, Grizzle WE, in: Bancroft JD, Gamble M (Eds.), Theory and Practice of Histological Techniques (Sixth Edition), Churchill Livingstone, Edinburgh, 2008, pp. 161–186. [Google Scholar]
- [21].Knudson CB, Knudson W, Semin. Cell Dev. Biol. 12 (2001) 69–78. [DOI] [PubMed] [Google Scholar]
- [22].Kiani C, Chen L, Wu YJ, Yee AJ, Yang BB, Cell Res. 12 (2002) 19–32. [DOI] [PubMed] [Google Scholar]
- [23].Solis MA, Chen Y-H, Wong TY, Bittencourt VZ, Lin Y-C, Huang LLH, Biochem. Res. Int. 2012 (2012) e346972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Slaughter BV, Khurshid SS, Fisher OZ, Khademhosseini A, Peppas NA, Adv. Mater. 21 (2009) 3307–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Walimbe T, Panitch A, Front. Pharmacol. 10 (2020) 1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Asgari M, Latifi N, Heris HK, Vali H, Mongeau L, Sci. Rep. 7 (2017) 1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Shoulders MD, Raines RT, Annu. Rev. Biochem. 78 (2009) 929–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bächinger HP, Mizuno K, Vranka JA, Boudko SP, in: Mander L, Liu H-W (Eds.), Comprehensive Natural Products II: Chemistry and Biology, 1st ed., Elsevier, Portland, OR, 2010, pp. 469–530. [Google Scholar]
- [29].Salvatore L, Gallo N, Natali ML, Campa L, Lunetti P, Madaghiele M, Blasi FS, Corallo A, Capobianco L, Sannino A, Mater. Sci. Eng. C 113 (2020) 110963. [DOI] [PubMed] [Google Scholar]
- [30].Kivirikko KI, in: Bittar EE, Bittar N (Eds.), Principles of Medical Biology, Elsevier, 1996, pp. 233–254. [Google Scholar]
- [31].Shaw LM, Olsen BR, Trends Biochem. Sci. 16 (1991) 191–194. [DOI] [PubMed] [Google Scholar]
- [32].Van Der Rest M, Garrone R, FASEB J. 5 (1991) 2814–2823. [PubMed] [Google Scholar]
- [33].Dhara S, Datta P, Pal P, Sarkar SD, in: Kim S-K (Ed.), Marine Proteins and Peptides, 1st ed., John Wiley & Sons, Ltd, Chichester, UK, 2013, pp. 589–629. [Google Scholar]
- [34].Kühn K, in: Mayne R, Burgeson RE (Eds.), Structure and Function of Collagen Types, Elsevier, 1987, pp. 1–42. [Google Scholar]
- [35].Rossert J, de Crombrugghe B, in: Bilezikian J, Raisz L, Rodan G (Eds.), Principles of Bone Biology, 2nd ed., Elsevier, 2002, pp. 189–210. [Google Scholar]
- [36].Sophia Fox AJ, Bedi A, Rodeo SA, Sports Health 1 (2009) 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Rani PU, Stringa E, Dharmavaram R, Chatterjee D, Tuan RS, Khillan JS, Dev. Dyn. 214 (1999) 26–43. [DOI] [PubMed] [Google Scholar]
- [38].Boudko SP, Engel J, Okuyama K, Mizuno K, Bächinger HP, Schumacher MA, J. Biol. Chem. 283 (2008) 32580–32589. [DOI] [PubMed] [Google Scholar]
- [39].Kuivaniemi H, Tromp G, Gene 707 (2019) 151–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Khoshnoodi J, Pedchenko V, Hudson BG, Microsc. Res. Tech. 71 (2008) 357–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Tanjore H, Kalluri R, Am. J. Pathol. 168 (2006) 715–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Leeming DJ, Karsdal MA, in: Biochemistry of Collagens, Laminins and Elastin, 2nd ed., Elsevier, 2019, pp. 51–57. [Google Scholar]
- [43].Bonod-Bidaud C, Roulet M, Hansen U, Elsheikh A, Malbouyres M, Ricard-Blum S, Faye C, Vaganay E, Rousselle P, Ruggiero F, J. Investig. Dermatol. 132 (2012) 1841–1849. [DOI] [PubMed] [Google Scholar]
- [44].Sun M, Chen S, Adams SM, Florer JB, Liu H, Kao WW-Y, Wenstrup RJ, Birk DE, J. Cell Sci. 124 (2011) 4096–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Sun S, Genovese F, Karsdal MA, in: Karsdal MA (Ed.), Biochemistry of Collagens, Laminins and Elastin, Elsevier, 2019, pp. 59–67. [Google Scholar]
- [46].Sun S, Karsdal MA, in: Karsdal MA (Ed.), Biochemistry of Collagens, Laminins and Elastin, Elsevier, 2016, pp. 49–55. [Google Scholar]
- [47].Sakai LY, Keene DR, Morris NP, Burgeson RE, J. Cell Biol. 103 (1986) 1577–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hansen NUB, Gudmann NS, Karsdal MA, in: Karsdal MA (Ed.), Biochemistry of Collagens, Laminins and Elastin, Elsevier, 2019, pp. 75–81. [Google Scholar]
- [49].Sibinga NES, Foster LC, Hsieh C-M, Perrella MA, Lee W-S, Endege WO, Sage EH, Lee M-E, Haber E, Circ. Res. 80 (1997) 532–541. [DOI] [PubMed] [Google Scholar]
- [50].Robertson AM, Watton PN, in: Becker S, Kuznetsov A (Eds.), Transport in Biological Media, 1st ed., Elsevier, 2013, pp. 275–347. [Google Scholar]
- [51].Martel-Pelletier J, Boileau C, Pelletier J-P, Roughley PJ, Best Pract. Res. Clin. Rheumatol. 22 (2008) 351–384. [DOI] [PubMed] [Google Scholar]
- [52].Blumbach K, Bastiaansen-Jenniskens YM, DeGroot J, Paulsson M, van Osch GJVM, Zaucke F, Arthritis Rheum. 60 (2009) 3676–3685. [DOI] [PubMed] [Google Scholar]
- [53].Eyre DR, Semin. Arthritis Rheum. 21 (1991) 2–11. [DOI] [PubMed] [Google Scholar]
- [54].Shen G, Orthod. Craniofac. Res. 8 (2005) 11–17. [DOI] [PubMed] [Google Scholar]
- [55].Kwan AP, Cummings CE, Chapman JA, Grant ME, J. Cell Biol. 114 (1991) 597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Luo YY, Szlarski PM, Kehlet SN, Karsdal MA, in: Karsdal MA (Ed.), Biochemistry of Collagens, Laminins and Elastin, Elsevier, 2019, pp. 99–106. [Google Scholar]
- [57].Blaschke UK, Eikenberry EF, Hulmes DJS, Galla H-J, Bruckner P, J. Biol. Chem. 275 (2000) 10370–10378. [DOI] [PubMed] [Google Scholar]
- [58].Lunstrum GP, Morris NP, McDonough AM, Keene DR, Burgeson RE, J. Cell Biol. 113 (1991) 963–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Chiquet M, Birk DE, Bönnemann CG, Koch M, Int. J. Biochem. Cell Biol. 53 (2014) 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Wälchli C, Koch M, Chiquet M, Odermatt BF, Trueb B, J. Cell Sci. 107 (1994) 669–681. [DOI] [PubMed] [Google Scholar]
- [61].Karimbux NY, Nishimura I, J. Dent. Res. 74 (1995) 313–318. [DOI] [PubMed] [Google Scholar]
- [62].Berthod F, Germain L, Guignard R, Lethias C, Garrone R, Damour O, van der Rest M, Auger FA, J. Investig. Dermatol. 108 (1997) 737–742. [DOI] [PubMed] [Google Scholar]
- [63].Anderson S, SundarRaj S, Fite D, Wessel H, SundarRaj N, Investig. Ophthamol. Vis. Sci. 41 (2000) 55–63. [PubMed] [Google Scholar]
- [64].Tzortzaki EG, Koutsopoulos AV, Dambaki KI, Lambiri I, Plataki M, Gordon MK, Gerecke DR, Siafakas NM, J. Histochem. Cytochem. 54 (2006) 693–700. [DOI] [PubMed] [Google Scholar]
- [65].Siebuhr AS, Karsdal MA, in: Karsdal MA (Ed.), Biochemistry of Collagens, Laminins and Elastin, Elsevier, 2016, pp. 87–91. [Google Scholar]
- [66].Ylönen R, Kyrönlahti T, Sund M, Ilves M, Lehenkari P, Tuukkanen J, Pihlajaniemi T, J. Bone Miner. Res. 20 (2005) 1381–1393. [DOI] [PubMed] [Google Scholar]
- [67].Härönen H, Zainul Z, Tu H, Naumenko N, Sormunen R, Miinalainen I, Shakirzyanova A, Oikarainen T, Abdullin A, Martin P, Santoleri S, Koistinaho J, Silman I, Giniatullin R, Fox MA, Heikkinen A, Pihlajaniemi T, Hum. Mol. Genet. 26 (2017) 2076–2090. [DOI] [PubMed] [Google Scholar]
- [68].Ansorge HL, Meng X, Zhang G, Veit G, Sun M, Klement JF, Beason DP, Soslowsky LJ, Koch M, Birk DE, J. Biol. Chem. 284 (2009) 8427–8438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Li D, Clark CC, Myers JC, J. Biol. Chem. 275 (2000) 22339–22347. [DOI] [PubMed] [Google Scholar]
- [70].Myers JC, Amenta PS, Dion AS, Sciancalepore JP, Nagaswami C, Weisel JW, Yurchenco PD, Biochem. J. 404 (2007) 535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Bretaud S, Guillon E, Karppinen S-M, Pihlajaniemi T, Ruggiero F, Matrix Biol. Plus 6–7 (2020) 100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Grässel S, Bauer RJ, Matrix Biol. 32 (2013) 64–73. [DOI] [PubMed] [Google Scholar]
- [73].Grässel S, Unsöld C, Schäcke H, Bruckner-Tuderman L, Bruckner P, Matrix Biol. 18 (1999) 309–317. [DOI] [PubMed] [Google Scholar]
- [74].Kassner A, Hansen U, Miosge N, Reinhardt DP, Aigner T, Bruckner-Tuderman L, Bruckner P, Grässel S, Matrix Biol. 22 (2003) 131–143. [DOI] [PubMed] [Google Scholar]
- [75].Kassner A, Tiedemann K, Notbohm H, Ludwig T, Mörgelin M, Reinhardt DP, Chu M-L, Bruckner P, Grässel S, J. Mol. Biol. 339 (2004) 835–853. [DOI] [PubMed] [Google Scholar]
- [76].Sand JMB, Karsdal MA, in: Karsdal MA (Ed.), Biochemistry of Collagens, Laminins and Elastin, Elsevier, 2016, pp. 101–106. [Google Scholar]
- [77].McGrath JA, Gatalica B, Christiano AM, si K, Owaribe K, McMillan JR, Eady RAJ, J. Uitto, Nat. Genet. 11 (1995) 83–86. [DOI] [PubMed] [Google Scholar]
- [78].Has C, Kern JS, Dermatol. Clin. 28 (2010) 61–66. [DOI] [PubMed] [Google Scholar]
- [79].Seppänen A, Autio-Harmainen H, Alafuzoff I, Särkioja T, Veijola J, Hurskainen T, Bruckner-Tuderman L, Tasanen K, Majamaa K, Matrix Biol. 25 (2006) 185–188. [DOI] [PubMed] [Google Scholar]
- [80].Franzke C-W, Bruckner P, Bruckner-Tuderman L, J. Biol. Chem. 280 (2005) 4005–4008. [DOI] [PubMed] [Google Scholar]
- [81].Watanabe M, Natsuga K, Nishie W, Kobayashi Y, Donati G, Suzuki S, Fujimura Y, Tsukiyama T, Ujiie H, Shinkuma S, Nakamura H, Murakami M, Ozaki M, Nagayama M, Watt FM, Shimizu H, ELife 6 (2017) e26635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Saarela J, Rehn M, Oikarinen A, Autio-Harmainen H, Pihlajaniemi T, Am. J. Pathol. 153 (1998) 611–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Fukai N, EMBO J. 21 (2002) 1535–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Utriainen A, Hum. Mol. Genet. 13 (2004) 2089–2099. [DOI] [PubMed] [Google Scholar]
- [85].Ricard-Blum S, Front. Biosci. 16 (2011) 674. [DOI] [PubMed] [Google Scholar]
- [86].Su J, Gorse K, Ramirez F, Fox MA, J. Comp. Neurol. 518 (2010) 229–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Sumiyoshi H, Mor N, Lee SY, Doty S, Henderson S, Tanaka S, Yoshioka H, Rattan S, Ramirez F, J. Cell Biol. 166 (2004) 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Koch M, Foley JE, Hahn R, Zhou P, Burgeson RE, Gerecke DR, Gordon MK, J. Biol. Chem. 276 (2001) 23120–23126. [DOI] [PubMed] [Google Scholar]
- [89].Willumsen N, Karsdal MA, in: Karsdal MA (Ed.), Biochemistry of Collagens, Laminins and Elastin, Elsevier, 2016, pp. 127–129. [Google Scholar]
- [90].Fitzgerald J, Bateman JF, FEBS Lett. 505 (2001) 275–280. [DOI] [PubMed] [Google Scholar]
- [91].Chou M-Y, Li H-C, Genom. 79 (2002) 395–401. [DOI] [PubMed] [Google Scholar]
- [92].Koch M, Schulze J, Hansen U, Ashwodt T, Keene DR, Brunken WJ, Burgeson RE, Bruckner P, Bruckner-Tuderman L, J. Biol. Chem. 279 (2004) 22514–22521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Kehlet SN, Karsdal MA, in: Karsdal MA (Ed.), Biochemistry of Collagens, Laminins and Elastin, Elsevier, 2016, pp. 135–137. [Google Scholar]
- [94].Koch M, Veit G, Stricker S, Bhatt P, Kutsch S, Zhou P, Reinders E, Hahn RA, Song R, Burgeson RE, Gerecke DR, Mundlos S, Gordon MK, J. Biol. Chem. 281 (2006) 21546–21557. [DOI] [PubMed] [Google Scholar]
- [95].Spivey KA, Chung I, Banyard J, Adini I, Feldman HA, Zetter BR, Oncog. 31 (2012) 2362–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Banyard J, Bao L, Hofer MD, Zurakowski D, Spivey KA, Feldman AS, Hutchinson LM, Kuefer R, Rubin MA, Zetter BR, Clin. Cancer Res. 13 (2007) 2634–2642. [DOI] [PubMed] [Google Scholar]
- [97].Koch M, Laub F, Zhou P, Hahn RA, Tanaka S, Burgeson RE, Gerecke DR, Ramirez F, Gordon MK, J. Biol. Chem. 278 (2003) 43236–43244. [DOI] [PubMed] [Google Scholar]
- [98].Matsuo N, Tanaka S, Yoshioka H, Koch M, Gordon MK, Ramirez F, Connect. Tissue Res. 49 (2008) 68–75. [DOI] [PubMed] [Google Scholar]
- [99].Parmar AS, Nunes AM, Baum J, Brodsky B, Biopolym. 97 (2012) 795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Hashimoto T, EMBO J. 21 (2002) 1524–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Shinwari JMA, Khan A, Awad S, Shinwari Z, Alaiya A, Alanazi M, Tahir A, Poizat C, Al Tassan N, Am. J. Hum. Genet. 96 (2015) 147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Kjeld NG, Karsdal MA, in: Karsdal MA (Ed.), Biochemistry of Collagens, Laminins and Elastin, Elsevier, 2016, pp. 147–150. [Google Scholar]
- [103].Sato K, Yomogida K, Wada T, Yorihuzi T, Nishimune Y, Hosokawa N, Nagata K, J. Biol. Chem. 277 (2002) 37678–37684. [DOI] [PubMed] [Google Scholar]
- [104].Plumb DA, Dhir V, Mironov A, Ferrara L, Poulsom R, Kadler KE, Thornton DJ, Briggs MD, Boot-Handford RP, J. Biol. Chem. 282 (2007) 12791–12795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Genovese F, Karsdal MA, in: Karsdal MA (Ed.), Biochemistry of Collagens, Laminins and Elastin, Elsevier, 2016, pp. 155–158. [Google Scholar]
- [106].Pace JM, Corrado M, Missero C, Byers PH, Matrix Biol. 22 (2003) 3–14. [DOI] [PubMed] [Google Scholar]
- [107].Hjorten R, Hansen U, Underwood RA, Telfer HE, Fernandes RJ, Krakow D, Sebald E, Wachsmann-Hogiu S, Bruckner P, Jacquet R, Landis WJ, Byers PH, Pace JM, Bone 41 (2007) 535–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Grimal S, Puech S, Wagener R, Ventéo S, Carroll P, Fichard-Carroll A, Glia 58 (2010) 1977–1987. [DOI] [PubMed] [Google Scholar]
- [109].Arvanitidis A, Karsdal MA, in: Karsdal MA (Ed.), Biochemistry of Collagens, Laminins and Elastin, Elsevier, 2016, pp. 159–161. [Google Scholar]
- [110].Veit G, Kobbe B, Keene DR, Paulsson M, Koch M, Wagener R, J. Biol. Chem. 281 (2006) 3494–3504. [DOI] [PubMed] [Google Scholar]
- [111].Bella J, Hulmes DJS, in: Parry DAD, Squire JM (Eds.), Fibrous Proteins: Structures and Mechanisms, Springer International Publishing, 2017. [Google Scholar]
- [112].Terzi A, Gallo N, Bettini S, Sibillano T, Altamura D, Madaghiele M, De Caro L, Valli L, Salvatore L, Sannino A, Giannini C, Macromol. Biosci. 20 (2020) 2000017. [DOI] [PubMed] [Google Scholar]
- [113].Kadler KE, Int. J. Exp. Path. 98 (2017) 4–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Fang M, Jacob R, McDougal O, Oxford JT, Protein Cell 3 (2012) 419–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Brodsky B, Persikov AV, in: Parry DAD, Squire JM (Eds.), Fibrous Proteins: Coiled-Coils, Collagen and Elastomers, Elsevier, 2005, pp. 301–339. [Google Scholar]
- [116].Yamauchi M, Sricholpech M, Essays Biochem. 52 (2012) 113–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Uitto J, Arch. Biochem. Biophys. 192 (1979) 371–379. [DOI] [PubMed] [Google Scholar]
- [118].Chung E, Keele EM, Miller EJ, Biochem. 13 (1974) 3459–3464. [DOI] [PubMed] [Google Scholar]
- [119].Albaugh VL, Mukherjee K, Barbul A, J. Nutr. (2017) 2011–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Ramshaw JAM, Shah NK, Brodsky B, J. Struct. Biol. 122 (1998) 86–91. [DOI] [PubMed] [Google Scholar]
- [121].Chow WY, Forman CJ, Bihan D, Puszkarska AM, Rajan R, Reid DG, Slatter DA, Colwell LJ, Wales DJ, Farndale RW, Duer MJ, Sci. Rep. 8 (2018) 13809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Persikov AV, Ramshaw JAM, Kirkpatrick A, Brodsky B, Biochem. 39 (2000) 14960–14967. [DOI] [PubMed] [Google Scholar]
- [123].Adzhubei AA, Sternberg MJE, Makarov AA, J. Mol. Biol. 425 (2013) 2100–2132. [DOI] [PubMed] [Google Scholar]
- [124].Luo T, Kiick KL, Eur. Polym. J. 49 (2013) 2998–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Kubyshkin V, Org. Biomol. Chem. 17 (2019) 8031–8047. [DOI] [PubMed] [Google Scholar]
- [126].Holmgren SK, Taylor KM, Bretscher LE, Raines RT, Nat. 392 (1998) 666–667. [DOI] [PubMed] [Google Scholar]
- [127].Holmgren SK, Bretscher LE, Taylor KM, Raines RT, Chem. Biol. 6 (1999) 63–70. [DOI] [PubMed] [Google Scholar]
- [128].Shoulders MD, Guzei IA, Raines RT, Biopolym. 89 (2008) 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Persikov AV, Ramshaw JAM, Kirkpatrick A, Brodsky B, J. Mol. Biol. 316 (2002) 385–394. [DOI] [PubMed] [Google Scholar]
- [130].Persikov AV, Ramshaw JAM, Kirkpatrick A, Brodsky B, Biochem. 44 (2005) 1414–1422. [DOI] [PubMed] [Google Scholar]
- [131].Krishna OD, Kiick KL, Biomacromolecules 10 (2009) 2626–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Kramer RZ, Venugopal MG, Bella J, Mayville P, Brodsky B, Berman HM, J. Mol. Biol. 301 (2000) 1191–1205. [DOI] [PubMed] [Google Scholar]
- [133].Fallas JA, Dong J, Tao YJ, Hartgerink JD, J. Biol. Chem. 287 (2012) 8039–8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Gauba V, Hartgerink JD, J. Am. Chem. Soc. 129 (2007) 2683–2690. [DOI] [PubMed] [Google Scholar]
- [135].Mohs A, Silva T, Yoshida T, Amin R, Lukomski S, Inouye M, Brodsky B, J. Biol. Chem. 282 (2007) 29757–29765. [DOI] [PubMed] [Google Scholar]
- [136].Lukomski S, Nakashima K, Abdi I, Cipriano VJ, Ireland RM, Reid SD, Adams GG, Musser JM, Infect. Immun. 68 (2000) 6542–6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Lukomski S, Nakashima K, Abdi I, Cipriano VJ, Shelvin BJ, Graviss EA, Musser JM, Infect. Immun. 69 (2001) 1729–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Bornstein P, Ann. Rev. Biochem. 43 (1974) 567–603. [DOI] [PubMed] [Google Scholar]
- [139].Orgel JPRO, Irving TC, Miller A, Wess TJ, Proc. Natl. Acad. Sci. U.S.A 103 (2006) 9001–9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Orgel JP, Wess TJ, Miller A, Structure 8 (2000) 137–142. [DOI] [PubMed] [Google Scholar]
- [141].Lin AC, Goh MC, Proteins 49 (2002) 378–384. [DOI] [PubMed] [Google Scholar]
- [142].Sasaki S, Ikeda T, Okihara S, Nishimura S, Nakadate R, Saeki H, Oki E, Mori M, Hashizume M, Maehara Y, Sci. Rep. 9 (2019) 9383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Von Der Mark K, in: Seibel M, Robins S, Bilezikian J (Eds.), Dynamics of Bone and Cartilage Metabolism, 2nd ed., Elsevier, 2006, pp. 3–40. [Google Scholar]
- [144].Adamczyk C, Milz S, Tischer T, Putz R, Benjamin M, J. Anat. 212 (2008) 645–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Line S, Rhodes C, Yamada Y, in: Noda M (Ed.), Cellular and Molecular Biology of Bone, Elsevier, 1993, pp. 539–555. [Google Scholar]
- [146].Bank RA, Beekman B, Tenni R, TeKoppele JM, Chromatogr J. B Biomed. Sci. Appl. 703 (1997) 267–272. [DOI] [PubMed] [Google Scholar]
- [147].Luo YY, Karsdal MA, in: Karsdal MA (Ed.), Biochemistry of Collagens, Laminins and Elastin, Elsevier, 2016, pp. 77–80. [Google Scholar]
- [148].Uitto J, Lichtenstein JR, J. Investig. Dermatol. 66 (1976) 59–79. [DOI] [PubMed] [Google Scholar]
- [149].Kaur J, Reinhardt DP, in: Vishwakarma A, Sharpe P, Shi S, Ramalingam M (Eds.), Stem Cell Biology and Tissue Engineering in Dental Sciences, 1st ed., Elsevier, 2015, pp. 25–45. [Google Scholar]
- [150].Bosman FT, Stamenkovic I, J. Pathol. 200 (2003) 423–428. [DOI] [PubMed] [Google Scholar]
- [151].Chattopadhyay S, Raines RT, Biopolym. 101 (2014) 821–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Kadler KE, Baldock C, Bella J, Boot-Handford RP, J. Cell Sci. 120 (2007) 1955–1958. [DOI] [PubMed] [Google Scholar]
- [153].Fraser JRE, Laurent TC, Laurent UBG, J. Intern. Med. 242 (1997) 27–33. [DOI] [PubMed] [Google Scholar]
- [154].Vasvani S, Kulkarni P, Rawtani D, Int. J. Biol. Macromol. 151 (2020) 1012–1029. [DOI] [PubMed] [Google Scholar]
- [155].Cowman MK, Lee H-G, Schwertfeger KL, McCarthy JB, Turley EA, Front. Immunol. 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Meyer K, Palmer J, J. Biol. Chem. 107 (1934) 629–634. [Google Scholar]
- [157].Balazs EA, Laurent TC, Jeanloz RW, Biochem. J. 235 (1986) 903–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Necas J, Bartosikova L, Brauner P, Kolar J, Vet. Med. 53 (2008) 397–411. [Google Scholar]
- [159].Kuo J-W, Prestwich GD, in: Ducheyne P (Ed.), Comprehensive Biomaterials, Elsevier, 2011, pp. 239–259. [Google Scholar]
- [160].Lapčík L, Lapčík L, De Smedt S, Demeester J, Chabreček P, Chem. Rev. 98 (1998) 2663–2684. [DOI] [PubMed] [Google Scholar]
- [161].Boeriu CG, Springer J, Kooy FK, van den Broek LAM, Eggink G, Int. J. Carbohydr. Chem. 2013 (2013) 1–14. [Google Scholar]
- [162].Chen WYJ, Abatangelo G, Wound Repair Regen. 7 (1999) 79–89. [DOI] [PubMed] [Google Scholar]
- [163].Morris ER, Rees DA, Welsh EJ, J. Mol. Biol. 138 (1980) 383–400. [DOI] [PubMed] [Google Scholar]
- [164].Essendoubi M, Gobinet C, Reynaud R, Angiboust JF, Manfait M, Piot O, Skin Res. Technol. 22 (2016) 55–62. [DOI] [PubMed] [Google Scholar]
- [165].Laurent TC, Laurent UB, Fraser JRE, Immunol. Cell Biol. 74 (1996) A1–A7. [DOI] [PubMed] [Google Scholar]
- [166].Gatej I, Popa M, Rinaudo M, Biomacromolecules 6 (2005) 61–67. [DOI] [PubMed] [Google Scholar]
- [167].Gupta RC, Lall R, Srivastava A, Sinha A, Front. Vet. Sci. 6 (2019) 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Tavianatou AG, Caon I, Franchi M, Piperigkou Z, Galesso D, Karamanos NK, FEBS J. 286 (2019) 2883–2908. [DOI] [PubMed] [Google Scholar]
- [169].Reitinger S, Lepperdinger G, Gerontol. 59 (2013) 71–76. [DOI] [PubMed] [Google Scholar]
- [170].Jiang Y, Wang H, Deng M, Wang Z, Zhang J, Wang H, Zhang H, Mater. Sci. Eng. C 59 (2016) 1038–1046. [DOI] [PubMed] [Google Scholar]
- [171].McKee CM, Penno MB, Cowman M, Burdick MD, Strieter RM, Bao C, Noble PW, J. Clin. Investig. 98 (1996) 2403–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].D’Agostino A, Stellavato A, Corsuto L, Diana P, Filosa R, La Gatta A, De Rosa M, Schiraldi C, Carbohydr. Polym. 157 (2017) 21–30. [DOI] [PubMed] [Google Scholar]
- [173].Kang L, Jia W, Li M, Wang Q, Wang C, Liu Y, Wang X, Jin L, Jiang J, Gu G, Chen Z, Carbohydr. Polym. 223 (2019) 115106. [DOI] [PubMed] [Google Scholar]
- [174].Wu M, Du Y, Liu Y, He Y, Yang C, Wang W, Gao F, PLoS One 9 (2014) e92857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Lees VC, West DC, in: Catravas JD, Callow AD, Ryan US (Eds.), Vascular Endothelium: Responses to Injury, Springer US, Boston, MA, 1996, pp. 336–336. [Google Scholar]
- [176].McCourt PAG, Matrix Biol. 18 (1999) 427–432. [DOI] [PubMed] [Google Scholar]
- [177].Tammi R, Ågren UM, Tuhkanen A-L, Tammi M, Hyaluronan Metabolism in Skin, Gustav Fischer Verlag/VCH Publishers, New York, NY, 1994. [DOI] [PubMed] [Google Scholar]
- [178].Tammi R, Säämänen A-M, Maibach H, Tammi M, J. Investig. Dermatol. 97 (1991) 126–130. [DOI] [PubMed] [Google Scholar]
- [179].Ragan C, Meyer K, J. Clin. Investig. 28 (1949) 56–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Balazs EA, in: Disorders of the Knee, 1974, pp. 63–75. [Google Scholar]
- [181].Lee HG, Cowman MK, Anal. Chem. 219 (1994) 278–287. [DOI] [PubMed] [Google Scholar]
- [182].Österlin S, Acta Opthalmol. 55 (1977) 353–361. [DOI] [PubMed] [Google Scholar]
- [183].Balazs EA, Denlinger JL, in: The Eye, 3rd ed., Elsevier, 1984, pp. 533–589. [Google Scholar]
- [184].Bishop P, Eye 10 (1996) 664–670. [DOI] [PubMed] [Google Scholar]
- [185].Bańkowski E, Sobolewski K, Chyczewski L, Jaworski S, Biol. Neonate 71 (1997) 11–21. [DOI] [PubMed] [Google Scholar]
- [186].Kanayama N, Goto J, Terao T, Pediatr. Res. 45 (1999) 510–514. [DOI] [PubMed] [Google Scholar]
- [187].Dahl L, Hopwood JJ, Laureni UBG, Lilja K, Tengblad A, Biochem. Med. 30 (1983) 280–283. [DOI] [PubMed] [Google Scholar]
- [188].Dahl LB, Dahl IMS, Børresen A-L, Biochem. Med. Metab. Biol. 35 (1986) 219–226. [DOI] [PubMed] [Google Scholar]
- [189].Laurent UBG, Exp. Eye Res. 33 (1981) 147–155. [DOI] [PubMed] [Google Scholar]
- [190].Laurent U, Törnquist P, Granath K, Lilja-Englind K, Ytterberg D, Acta Opthalmol. 68 (1990) 109–112. [Google Scholar]
- [191].Tengblad A, Laurent UBG, Lilja K, Cahill RNP, Engström-Laurent A, Fraser JRR, Hansson HE, Laurent TC, Biochem. J. 236 (1986) 521–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Hill DR, Rho HK, Kessler SP, Amin R, Homer CR, McDonald C, Cowman MK, J. Biol. Chem. 288 (2013) 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Yuan T, He L, Yang J, Zhang L, Xiao Y, Fan Y, Zhang X, Process Biochem. 50 (2015) 2242–2250. [Google Scholar]
- [194].Sasaki Y, Uzuki M, Nohmi K, Kitagawa H, Kamataki A, Komagamine M, Murakami K, Sawai T, Int. J. Rheum. Dis. 14 (2011) 313–319. [DOI] [PubMed] [Google Scholar]
- [195].Weigel PH, in: Garg HG, Hales CA (Eds.), Chemistry and Biology of Hyaluronan, Elsevier, 2004, pp. 553–567. [Google Scholar]
- [196].Jokela TA, Jauhiainen M, Auriola S, Kauhanen M, Tiihonen R, Tammi MI, Tammi RH, J. Biol. Chem. 283 (2008) 7666–7673. [DOI] [PubMed] [Google Scholar]
- [197].Chao H, Spicer AP, J. Biol. Chem. 280 (2005) 27513–27522. [DOI] [PubMed] [Google Scholar]
- [198].Noble PW, Matrix Biol. 21 (2002) 25–29. [DOI] [PubMed] [Google Scholar]
- [199].Bastow ER, Byers S, Golub SB, Clarkin CE, Pitsillides AA, Fosang AJ, Cell. Mol. Life Sci. 65 (2008) 395–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Stern R, Kogan G, Jedrzejas MJ, Šoltés L, Biotechnol. Adv. 25 (2007) 537–557. [DOI] [PubMed] [Google Scholar]
- [201].Girish KS, Kemparaju K, Life Sci. 80 (2007) 1921–1943. [DOI] [PubMed] [Google Scholar]
- [202].Krupkova O, Greutert H, Boos N, Lemcke J, Liebscher T, Wuertz-Kozak K, Eur. Spine J. 29 (2020) 605–615. [DOI] [PubMed] [Google Scholar]
- [203].Cyphert JM, Trempus CS, Garantziotis S, Int. J. Cell Biol. 2015 (2015) 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Stern R, Jedrzejas MJ, Chem. Rev. 106 (2006) 818–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].El-Safory NS, Fazary AE, Lee C-K, Carbohydr. Polym. 81 (2010) 165–181. [Google Scholar]
- [206].Knudson CB, Birth Defects Res. C Embryo Today Rev. 69 (2003) 174–196. [DOI] [PubMed] [Google Scholar]
- [207].Lebbink RJ, de Ruiter T, Adelmeijer J, Brenkman AB, van Helvoort JM, Koch M, Farndale RW, Lisman T, Sonnenberg A, Lenting PJ, Meyaard L, J. Exp. Med. 203 (2006) 1419–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Goldring MB, Goldring SR, J. Cell. Physiol. 213 (2007) 626–634. [DOI] [PubMed] [Google Scholar]
- [209].Humphries JD, Humphries MJ, J. Cell Sci. 119 (2006) 3901–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Heino J, Bioessays 29 (2007) 1001–1010. [DOI] [PubMed] [Google Scholar]
- [211].Barczyk M, Carracedo S, Gullberg D, Cell Tissue Res. 339 (2010) 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Heino J, Huhtala M, Käpylä J, Johnson MS, Int. J. Biochem. (2009) 341–348. [DOI] [PubMed] [Google Scholar]
- [213].Nyberg P, Xie L, Kalluri R, Cancer Res. 65 (2005) 3967–3979. [DOI] [PubMed] [Google Scholar]
- [214].Ortega N, J. Cell Sci. 115 (2002) 4201–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Basakran NS, Saudi Med. J. 36 (2015) 273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Ghosh P, Guidolin D, Semin. Arthritis Rheum. 32 (2002) 10–37. [DOI] [PubMed] [Google Scholar]
- [217].Turley EA, Noble PW, Bourguignon LYW, J. Biol. Chem. 277 (2002) 4589–4592. [DOI] [PubMed] [Google Scholar]
- [218].Bhattacharya DS, Svechkarev D, Souchek JJ, Hill TK, Taylor MA, Natarajan A, Mohs AM, J. Mater. Chem. B 5 (2017) 8183–8192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Yang B, Yang BL, Savani RC, Turley EA, EMBO JF 13 (1994) 286–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Camenisch TD, McDonald JA, Am. J. Respir. Cell Mol. Biol. 23 (2000) 431–433. [DOI] [PubMed] [Google Scholar]
- [221].Naahidi S, Jafari M, Logan M, Wang Y, Yuan Y, Bae H, Dixon B, Chen P, Biotechnol. Adv. 35 (2017) 530–544. [DOI] [PubMed] [Google Scholar]
- [222].Toole BP, Semin. Cell Dev. Biol. 12 (2001) 79–87. [DOI] [PubMed] [Google Scholar]
- [223].Huang L, Yoneda M, Kimata K, J. Biol. Chem. 268 (1993) 26725–26730. [PubMed] [Google Scholar]
- [224].Qin J, Luo T, Kiick KL, Biomacromolecules 20 (2019) 1514–1521. [DOI] [PubMed] [Google Scholar]
- [225].Sun J, Zhang Y, Li B, Gu Y, Chen L, J. Mater. Chem. B 5 (2017) 8770–8779. [DOI] [PubMed] [Google Scholar]
- [226].Thapa RK, Kiick KL, Sullivan MO, Acta Biomater. 103 (2020) 115–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Sun X, Liu Z, Zhao S, Xu X, Wang S, Guo C, Xiao J, J. Mater. Chem. B 7 (2019) 3201–3209. [Google Scholar]
- [228].Cosgriff-Hernandez E, Hahn MS, Russell B, Wilems T, Munoz-Pinto D, Browning MB, Rivera J, Höök M, Acta Biomater. 6 (2010) 3969–3977. [DOI] [PubMed] [Google Scholar]
- [229].Peng YY, Yoshizumi A, Danon SJ, Glattauer V, Prokopenko O, Mirochnitchenko O, Yu Z, Inouye M, Werkmeister JA, Brodsky B, Ramshaw JAM, Biomater. 31 (2010) 2755–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Zhang J, Ma X, Fan D, Zhu C, Deng J, Hui J, Ma P, Mater. Sci. Eng. C 43 (2014) 547–554. [DOI] [PubMed] [Google Scholar]
- [231].Seo N, Russell BH, Rivera JJ, Liang X, Xu X, Afshar-Kharghan V, Höök M, J. Biol. Chem. 285 (2010) 31046–31054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Werkmeister JA, Ramshaw JAM, Biomed. Mater. 7 (2012) 012002. [DOI] [PubMed] [Google Scholar]
- [233].León-López A, Morales-Peñaloza A, Martínez-Juárez VM, Vargas-Torres A, Zeugolis DI, Aguirre-Álvarez G, Mol. 24 (2019) 4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Fietzek PP, Kühn K, Hall DA, Jackson DS, in: International Review of Connective Tissue Research, Elsevier, 1976, pp. 1–60. [DOI] [PubMed] [Google Scholar]
- [235].Boot-Handford RP, Tuckwell DS, Bioessays 25 (2003) 142–151. [DOI] [PubMed] [Google Scholar]
- [236].Eastoe JE, Biochem. J. 61 (1955) 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Davison-Kotler E, Marshall WS, García-Gareta E, Bioeng. 6 (2019) 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Ferraro V, Gaillard-Martinie B, Sayd T, Chambon C, Anton M, Santé-Lhoutellier V, Int. J. Biol. Macromol. 97 (2017) 55–66. [DOI] [PubMed] [Google Scholar]
- [239].Avila Rodríguez MI, Rodríguez Barroso LG, Sánchez ML, J. Cosmet. Dermatol. 17 (2018) 20–26. [DOI] [PubMed] [Google Scholar]
- [240].Johnson K, Biomater. 20 (1999) 1003–1015. [DOI] [PubMed] [Google Scholar]
- [241].Wood A, Ogawa M, Portier RJ, Schexnayder M, Shirley M, Losso JN, Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 151 (2008) 246–249. [DOI] [PubMed] [Google Scholar]
- [242].Kumar KVSS, Sai KP, Babu M, J. Biomed. Mater. Res. 61 (2002) 197–202. [DOI] [PubMed] [Google Scholar]
- [243].Wolf K, Alexander S, Schacht V, Coussens LM, von Andrian UH, van Rheenen J, Deryugina E, Friedl P, Semin. Cell Dev. Biol. 20 (2009) 931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [244].Schmidt MM, Dornelles RCP, Mello RO, Kubota EH, Mazutti MA, Kempka AP, Demiate IM, Int. Food Res. J. 23 (2016) 913–922. [Google Scholar]
- [245].Kreger S, Bell B, Bailey J, Stites E, Kuske J, Waisner B, Voytik- Harbin S, Biopolym. Orig. Res. Biomol. 93 (2010) 690–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Ghodbane SA, Dunn MG, J. Biomed. Mater. Res. 104 (2016) 2685–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Parenteau-Bareil R, Gauvin R, Cliche S, Gariépy C, Germain L, Berthod F, Acta Biomater. 7 (2011) 3757–3765. [DOI] [PubMed] [Google Scholar]
- [248].Egorikhina MN, Aleynik DY, Rubtsova YP, Levin GY, Charykova IN, Semenycheva LL, Bugrova ML, Zakharychev EA, Bioact. Mater. 4 (2019) 334–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [249].Park S-H, Song T, Bae TS, Khang G, Choi BH, Park SR, Min B-H, Int. J. Precis. Eng. Manuf. 13 (2012) 2059–2066. [Google Scholar]
- [250].Damodarasamy M, Vernon RB, Karres N, Chang CH, Bianchi-Frias D, Nelson PS, Reed MJ, J. Gerontol. Ser. A Biomed. Sci. Med. Sci. 65 (2010) 209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [251].Lynn AK, Yannas IV, Bonfield W, J. Biomed. Mater. Res. 71B (2004) 343–354. [DOI] [PubMed] [Google Scholar]
- [252].Cooperman L, Michaeli D, J. Am. Acad. Dermatol. 10 (1984) 638–646. [DOI] [PubMed] [Google Scholar]
- [253].Cooperman L, Michaeli D, J. Am. Acad. Dermatol. 10 (1984) 647–651. [DOI] [PubMed] [Google Scholar]
- [254].Charriere G, Bejot M, Schnitzler L, Ville G, Hartmann DJ, J. Am. Acad. Dermatol. 21 (1989) 1203–1208. [DOI] [PubMed] [Google Scholar]
- [255].Eaglstein WH, Alvarez OM, Auletta M, Leffel D, Rogers GS, Zitelli JA, Norris JEC, Thomas I, Irondo M, Fewkes J, Hardin-Young J, Duff RG, Sabolinski ML, Dermatol. Surg. 25 (1999) 195–201. [DOI] [PubMed] [Google Scholar]
- [256].Furthmayr H, Timpl R, in: Hall DA, Jackson DS (Eds.), International Review of Connective Tissue Research, Elsevier, 1976, pp. 61–99. [DOI] [PubMed] [Google Scholar]
- [257].Helseth DL, Veis A, J. Biol. Chem. 256 (1981) 7118–7128. [PubMed] [Google Scholar]
- [258].Brennan M, Davison PF, Biopolym. 20 (1981) 2195–2202. [DOI] [PubMed] [Google Scholar]
- [259].Trentham DE, Townes AS, Kang AH, J. Exp. Med. 146 (1977) 857–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [260].Svensson L, Jirholt, Holmdahl R, Jansson L, Clin. Exp. Immunol. 111 (1998) 521–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Courtenay JS, Dallman MJ, Dayan AD, Martin A, Mosedale B, Nat. 283 (1980) 666–668. [DOI] [PubMed] [Google Scholar]
- [262].Berillis P, in: Research Trends in Biochemistry, Molecular Biology and Microbiology, 2015, pp. 1–13. [Google Scholar]
- [263].Melander RJ, Liu H, Stephens MD, Bewley CA, Melander C, Bioorganic Med. Chem. Lett. 26 (2016) 5863–5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [264].Roulet M, Välkkilä M, Chanut-Delalande H, Hämäläinen E-R, Kessler E, Ala-Kokko L, Männikkö M, Bonod-Bidaud C, Ruggiero F, J. Biomed. Biotechnol. 2010 (2010) 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [265].Shi J, Ma X, Gao Y, Fan D, Zhu C, Mi Y, Xue W, Protein J. 36 (2017) 322–331. [DOI] [PubMed] [Google Scholar]
- [266].Rutschmann C, Baumann S, Cabalzar J, Luther KB, Hennet T, Appl. Microbiol. Biotechnol. 98 (2014) 4445–4455. [DOI] [PubMed] [Google Scholar]
- [267].Toman PD, Chisholm G, McMullin H, Giere LM, Olsen DR, Kovach RJ, Leigh SD, Fong BE, Chang R, Daniels GA, Berg RA, Hitzeman RA, J. Biol. Chem. 275 (2000) 23303–23309. [DOI] [PubMed] [Google Scholar]
- [268].Vaughn PR, Galanis M, Richards KM, Tebb TA, Ramshaw JA, Werkmeister JA, DNA Cell Biol. 17 (1998) 511–518. [DOI] [PubMed] [Google Scholar]
- [269].Chan SWP, Hung S-P, Raman SK, Hatfield GW, Lathrop RH, Da Silva NA, Wang S-W, Biomacromolecules 11 (2010) 1460–1469. [DOI] [PubMed] [Google Scholar]
- [270].Olsen DR, Leigh SD, Chang R, McMullin H, Ong W, Tai E, Chisholm G, Birk DE, Berg RA, Hitzeman RA, Toman PD, J. Biol. Chem. 276 (2001) 24038–24043. [DOI] [PubMed] [Google Scholar]
- [271].Nokelainen M, Tu H, Vuorela A, Notbohm H, Kivirikko KI, Myllyharju J, Yeast 18 (2001) 797–806. [DOI] [PubMed] [Google Scholar]
- [272].Ruottinen M, Bollok M, Kogler M, Neubauer A, Krause M, Hamalainen E-R, Myllyharju J, Vasala A, Neubauer P, BMC Biotechnol. 8 (2008) 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [273].Wang L, Fan D, He J, Lv Z, Zhu C, Biotechnol. Bioprocess Eng. 19 (2014) 916–924. [Google Scholar]
- [274].Vuorela A, EMBO J. 16 (1997) 6702–6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [275].Ruggiero F, Exposito J-Y, Bournat P, Gruber V, Perret S, Comte J, Olagnier B, Garrone R, Theisen M, FEBS Lett. 469 (2000) 132–136. [DOI] [PubMed] [Google Scholar]
- [276].Stein H, Wilensky M, Tsafrir Y, Rosenthal M, Amir R, Avraham T, Ofir K, Dgany O, Yayon A, Shoseyov O, Biomacromolecules 10 (2009) 2640–2645. [DOI] [PubMed] [Google Scholar]
- [277].Merle C, Perret S, Lacour T, Jonval V, Hudaverdian S, Garrone R, Ruggiero F, Theisen M, FEBS Lett. 515 (2002) 114–118. [DOI] [PubMed] [Google Scholar]
- [278].Xu X, Gan Q, Clough RC, Pappu KM, Howard JA, Baez JA, Wang K, BMC Biotechnol. 11 (2011) 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [279].Fukuda K, Hori H, Utani A, Burbelo PD, Yamada Y, Biochem. Biophys. Res. Commun. 231 (1997) 178–182. [DOI] [PubMed] [Google Scholar]
- [280].Mazzorana M, Gruffat H, Sergeant A, van der Rest M, J. Biol. Chem. 268 (1993) 3029–3032. [PubMed] [Google Scholar]
- [281].Fichard A, Tillet E, Delacoux F, Garrone R, Ruggiero F, J. Biol. Chem. 272 (1997) 30083–30087. [DOI] [PubMed] [Google Scholar]
- [282].Chen M, Costa FK, Lindvay CR, Han Y-P, Woodley DT, J. Biol. Chem. 277 (2002) 2118–2124. [DOI] [PubMed] [Google Scholar]
- [283].Frischholz S, Beier F, Girkontaite I, Wagner K, Pöschl E, Turnay J, Mayer U, Von K, J. Biol. Chem. 273 (1998) 4547–4555. [DOI] [PubMed] [Google Scholar]
- [284].Geddis AE, Prockop DJ, Matrix 13 (1993) 399–405. [DOI] [PubMed] [Google Scholar]
- [285].Wieczorek A, Rezaei N, Chan CK, Xu C, Panwar P, Brömme D, Merschrod S. EF, Forde NR, BMC Biotechnol. 15 (2015) 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [286].Myllyharju J, Lamberg A, Notbohm H, Fietzek PP, Pihlajaniemi T, Kivirikko KI, J. Biol. Chem. 272 (1997) 21824–21830. [DOI] [PubMed] [Google Scholar]
- [287].Tomita M, Ohkura N, Ito M, Kato T, Royce PM, Kitajima T, Biochem. J. 312 (1995) 847–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [288].Lamberg A, Helaakoski T, Myllyharju J, Peltonen S, Notbohm H, Pihlajaniemi T, Kivirikko KI, J. Biol. Chem. 271 (1996) 11988–11995. [DOI] [PubMed] [Google Scholar]
- [289].Pihlajamaa T, Perälä M, Vuoristo MM, Nokelainen M, Bodo M, Schulthess T, Vuorio E, Timpl R, Engel J, Ala-Kokko L, J. Biol. Chem. 274 (1999) 22464–22468. [DOI] [PubMed] [Google Scholar]
- [290].Qi Q, Yao L, Liang Z, Yan D, Li Z, Huang Y, Sun J, Mol. Genet. Genom. 291 (2016) 2189–2198. [DOI] [PubMed] [Google Scholar]
- [291].Li H-C, Huang C-C, Chen S-F, Chou M-Y, Biochem. Biophys. Res. Commun. 336 (2005) 375–385. [DOI] [PubMed] [Google Scholar]
- [292].Adachi T, Wang X, Murata T, Obara M, Akutsu H, Machida M, Umezawa A, Tomita M, Biotechnol. Bioeng. 106 (2010) 860–870. [DOI] [PubMed] [Google Scholar]
- [293].Tomita M, Munetsuna H, Sato T, Adachi T, Hino R, Hayashi M, Shimizu K, Nakamura N, Tamura T, Yoshizato K, Nat. Biotechnol. 21 (2003) 52–56. [DOI] [PubMed] [Google Scholar]
- [294].Cheng Y, Shi W, Xiao X, Zhang Q, Zhang Q, Su Z, Xiang Q, Huang Y, Pak. J. Zool. 52 (2019) 321–330. [Google Scholar]
- [295].Ruggiero F, Koch M, Methods 45 (2008) 75–85. [DOI] [PubMed] [Google Scholar]
- [296].Jiang X, Wang Y, Fan D, Zhu C, Liu L, Duan Z, J. Biomater. Appl. 31 (2017) 1099–1107. [DOI] [PubMed] [Google Scholar]
- [297].Hu K, Lv Q, Cui FZ, Feng QL, Kong XD, Wang HL, Huang LY, Li T, J. Bioact. Compat. Polym. 21 (2006) 23–37. [Google Scholar]
- [298].Pulkkinen HJ, Tiitu V, Valonen P, Hämäläinen E-R, Lammi MJ, Kiviranta I, Int. J. Artif. Organs 31 (2008) 960–969. [DOI] [PubMed] [Google Scholar]
- [299].Pulkkinen HJ, Tiitu V, Valonen P, Jurvelin JS, Lammi MJ, Kiviranta I, Osteoarthr. Cartil. 18 (2010) 1077–1087. [DOI] [PubMed] [Google Scholar]
- [300].He Y, Hou Z, Wang J, Wang Z, Li X, Liu J, XiaolinYang, Liang Q, Zhao J, Int. J. Biol. Macromol. 149 (2020) 1275–1284. [DOI] [PubMed] [Google Scholar]
- [301].Fakhari A, Berkland C, Acta Biomater. 9 (2013) 7081–7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [302].In: Hyaluronic Acid, John Wiley & Sons, Ltd, Chichester, UK, 2015, pp. 77–95. [Google Scholar]
- [303].Schiraldi C, Andreozzi L, Marzaioli I, Vinciguerra S, D’Avino A, Volpe F, Panariello A, De Rosa M, Biocatal. Biotransformation 28 (2010) 83–89. [Google Scholar]
- [304].de Melo BAG, Santana MHA, Appl. Biochem. Biotechnol. 189 (2019) 424–436. [DOI] [PubMed] [Google Scholar]
- [305].Cardoso MJ, Caridade SG, Costa RR, Mano JF, Biomacromolecules 17 (2016) 1347–1357. [DOI] [PubMed] [Google Scholar]
- [306].Hascall V, Esko JD, in: Varki A, Cummings RD, Esko J (Eds.), Essentials of Glycobiology, 2nd ed., Cold Spring Harbor Laboratory Press, New York, NY, 2009. [PubMed] [Google Scholar]
- [307].Sze JH, Brownlie JC, Love CA, 3 Biotech 6 (2016) 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [308].Balazs EA, Riverdale NY, US 4141973A - Ultrapure Hyaluronic Acid And The Use Thereof The Lens, 1979.
- [309].Allison DD, Grande-Allen KJ, Tissue Eng. 12 (2006) 2131–2140. [DOI] [PubMed] [Google Scholar]
- [310].Kogan G, Šoltés L, Stern R, Gemeiner P, Biotechnol. Lett. 29 (2006) 17–25. [DOI] [PubMed] [Google Scholar]
- [311].DeAngelis PL, Papaconstantinouan J, Weigel PH, J. Biol. Chem. 268 (1993) 14568–14571. [PubMed] [Google Scholar]
- [312].DeAngelis PL, Cell. Mol. Life Sci. 56 (1999) 670–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [313].Pan NC, Pereira HCB, da Silva M. de L.C, Vasconcelos AFD, Celligoi MAPC, Appl. Biochem. Biotechnol. 182 (2017) 276–293. [DOI] [PubMed] [Google Scholar]
- [314].Zakeri A, Rasaee MJ, Pourzardosht N, Biotechnol. Rep. 16 (2017) 65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [315].Pourzardosht N, Rasaee MJ, Mol. Biotechnol. 59 (2017) 192–199. [DOI] [PubMed] [Google Scholar]
- [316].Rohit SG, Jyoti PK, Subbi RRT, Naresh M, Senthilkumar S, Biochem. Eng. J. 137 (2018) 284–293. [Google Scholar]
- [317].Attia YA, Kobeasy MI, Samer M, Carbohydr. Polym. 192 (2018) 135–142. [DOI] [PubMed] [Google Scholar]
- [318].Mohan N, Pavan SS, Achar A, Swaminathan N, Sivaprakasam S, Biochem. Eng. J. 152 (2019) 107367. [Google Scholar]
- [319].Cavalcanti ADD, Melo BAG, Oliveira RC, Santana MHA, Appl. Biochem. Biotechnol. 188 (2019) 527–539. [DOI] [PubMed] [Google Scholar]
- [320].Güngör G, Gedikli S, Toptaş Y, Akgün DE, Demirbilek M, Yazıhan N, Aytar Çelik P, Denkbaş EB, Çabuk A, J. Chem. Technol. Biotechnol. 94 (2019) 1843–1852. [Google Scholar]
- [321].Chen WY, Marcellin E, Hung J, Nielsen LK, J. Biol. Chem. 284 (2009) 18007–18014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [322].Jia Y, Zhu J, Chen X, Tang D, Su D, Yao W, Gao X, Bioresour. Technol. 132 (2013) 427–431. [DOI] [PubMed] [Google Scholar]
- [323].Li Y, Li G, Zhao X, Shao Y, Wu M, Ma T , 3 Biotech 9 (2019) 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [324].Jeeva P, Shanmuga Doss S, Sundaram V, Jayaraman G, Appl. Microbiol. Biotechnol. 103 (2019) 4363–4375. [DOI] [PubMed] [Google Scholar]
- [325].Westbrook AW, Ren X, Moo-Young M, Chou CP, Biotechnol. Bioeng. 115 (2018) 1239–1252. [DOI] [PubMed] [Google Scholar]
- [326].Woo JE, Seong HJ, Lee SY, Jang Y-S, Front. Bioeng. Biotechnol. 7 (2019) 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [327].Mao Z, Shin H-D, Chen R, Appl. Microbiol. Biotechnol. 84 (2009) 63–69. [DOI] [PubMed] [Google Scholar]
- [328].Yu H, Stephanopoulos G, Metab. Eng. 10 (2008) 24–32. [DOI] [PubMed] [Google Scholar]
- [329].Wang Y, Hu L, Huang H, Wang H, Zhang T, Chen J, Du G, Kang Z, Nat. Commun. 11 (2020) 3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [330].Zhang L, Toscano Selão T, Nixon PJ, Norling B, Algal Res. 44 (2019) 101702. [Google Scholar]
- [331].Gomes AMV, Netto JHCM, Carvalho LS, Parachin NS, Microorg. 7 (2019) 294. [Google Scholar]
- [332].Chahuki FF, Aminzadeh S, Jafarian V, Tabandeh F, Khodabandeh M, Int. J. Biol. Macromol. 121 (2019) 870–881. [DOI] [PubMed] [Google Scholar]
- [333].Cheng F, Yu H, Stephanopoulos G, Metab. Eng. 55 (2019) 276–289. [DOI] [PubMed] [Google Scholar]
- [334].Marcellin E, Steen JA, Nielsen LK, Appl. Microbiol. Biotechnol. 98 (2014) 6947–6956. [DOI] [PubMed] [Google Scholar]
- [335].Chien L-J, Lee C-K, Biotechnol. Prog. 23 (2007) 1017–1022. [DOI] [PubMed] [Google Scholar]
- [336].Widner B, Behr R, Von Dollen S, Tang M, Heu T, Sloma A, Sternberg D, DeAngelis PL, Weigel PH, Brown S, Appl. Environ. Microbiol. 71 (2005) 3747–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [337].Hmar RV, Prasad SB, Jayaraman G, Ramachandran KB, Biotechnol. J. 9 (2014) 1554–1564. [DOI] [PubMed] [Google Scholar]
- [338].Chien L-J, Lee C-K, Appl. Microbiol. Biotechnol. 77 (2007) 339–346. [DOI] [PubMed] [Google Scholar]
- [339].Liu L, Liu Y, Li J, Du G, Chen J, Microb. Cell Fact. 10 (2011) 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [340].Schulte S, Doss SS, Jeeva P, Ananth M, Blank LM, Jayaraman G, Appl. Biochem. Biotechnol. 103 (2019) 7567–7581. [DOI] [PubMed] [Google Scholar]
- [341].Chen WY, Marcellin E, Steen JA, Nielsen LK, Mol. Biotechnol. 56 (2014) 147–156. [DOI] [PubMed] [Google Scholar]
- [342].Jing W, DeAngelis PL, Glycobiology 10 (2000) 883–889. [DOI] [PubMed] [Google Scholar]
- [343].Mandawe J, Infanzon B, Eisele A, Zaun H, Kuballa J, Davari MD, Jakob F, Elling L, Schwaneberg U, Chembiochem 19 (2018) 1414–1423. [DOI] [PubMed] [Google Scholar]
- [344].Jing W, DeAngelis PL, J. Biol. Chem. 279 (2004) 42345–42349. [DOI] [PubMed] [Google Scholar]
- [345].Agarwal G, Prasad KKV,SB, Bhaduri A, Jayaraman G, Sci. Rep. 9 (2019) 12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [346].Chvapil M, J. Biomed. Mater. Res. 11 (1977) 721–741. [DOI] [PubMed] [Google Scholar]
- [347].Böttcher-Haberzeth S, Biedermann T, Schiestl C, Hartmann-Fritsch F, Schneider J, Reichmann E, Meuli M, Pediatr. Surg. Int. 28 (2012) 171–177. [DOI] [PubMed] [Google Scholar]
- [348].Mattern R-H, Pierschbacher MD, Cahn F, Tschopp JF, Malaney TI, Collagen/Glycosaminoglycan Matrix Stable to Sterilizing by Electron Beam Radiation, US6969523B1, 2005.
- [349].Cervelli V, Brinci L, Spallone D, Tati E, Palla L, Lucarini L, Angelis BD, Int. Wound. J. 8 (2011) 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [350].Malessa R, Kassner A, Degradation-Stabilised, Biocompatible Collagen Matrices, US9327033B2, 2016.
- [351].Curran MP, Plosker GL, BioDrugs 16 (2002) 439–455. [DOI] [PubMed] [Google Scholar]
- [352].Eaglstein WH, Falanga V, Clin. Ther. 19 (1997) 894–905. [DOI] [PubMed] [Google Scholar]
- [353].Kemp P, Bell E, Kagan DT, Mason V, Cavallaro J, Preparation of Tissue Equivalents by Contraction of a Collagen Gel Layered on a Collagen Gel, US5536656A, 1996.
- [354].Glassman SD, Carreon L, Djurasovic M, Campbell MJ, Puno RM, Johnson JR, Dimar JR, Spine J. 7 (2007) 44–49. [DOI] [PubMed] [Google Scholar]
- [355].McKay WF, Peckham SM, Badura JM, Int. Othop. 31 (2007) 729–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [356].Bai X, Gao M, Syed S, Zhuang J, Xu X, Zhang X-Q, Bioact. Mater. 3 (2018) 401–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [357].Epstein NE, Surg. Neurol. Int. 2 (2011) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [358].Donaldson J, Tudor F, McDermott ID, Surg. Neurol. Int. 29 (2015) 24–30. [Google Scholar]
- [359].Vinatier C, Guicheux J, Ann. Phys. Rehabil. Med. 59 (2016) 139–144. [DOI] [PubMed] [Google Scholar]
- [360].Debus ES, Geiger D, Sailer M, Ederer J, Thiede A, Eur. Surg. Res. 29 (1997) 52–61. [DOI] [PubMed] [Google Scholar]
- [361].Pavan A, Bosio M, Longo T, J. Biomed. Mater. Res. 13 (1979) 477–496. [DOI] [PubMed] [Google Scholar]
- [362].McLaughlin S, McNeill B, Podrebarac J, Hosoyama K, Sedlakova V, Cron G, Smyth D, Seymour R, Goel K, Liang W, Rayner KJ, Ruel M, Suuronen EJ, Alarcon EI, Nat. Commun. 10 (2019) 4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [363].Muhonen V, Narcisi R, Nystedt J, Korhonen M, van Osch GJVM, Kiviranta I, J. Tissue. Eng. Regen. Med. 11 (2015) 843–854. [DOI] [PubMed] [Google Scholar]
- [364].Haagdorens M, Cėpla V, Melsbach E, Koivusalo L, Skottman H, Griffith M, Valiokas R, Zakaria N, Pintelon I, Tassignon M-J, Stem Cells Int. 2019 (2019) 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [365].Muraya K, Kawasaki T, Yamamoto T, Akutsu H, BioResearch Open Access 8 (2019) 210–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [366].Ramírez-Rodríguez GB, Montesi M, Panseri S, Sprio S, Tampieri A, Sandri M, Tissue Engineering Part A 23 (2017) 1423–1435. [DOI] [PubMed] [Google Scholar]
- [367].Que RA, Arulmoli J, Da Silva NA, Flanagan LA, Wang S-W, J. Biomed. Mater. Res. Part A 106A (2018) 1363–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [368].Smit NW, ten Sande JN, Parvizi M, van Amersfoorth SCM, Plantinga JA, van Spreuwel-Goossens CAFM, van Dongen EMWM, van Dessel PFHM, Kluijtmans SGJM, Meijborg VMF, de Bakker JMT, Harmsen MC, Coronel R, PLOS ONE 12 (2017) 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [369].Tsukioka T, Hiratsuka T, Nakamura M, Watanabe T, Kitamura Y, Isobe K, Okudera T, Okudera H, Azuma A, Uematsu K, Nakata K, Kawase T, J. Biomed. Mater. Res B. 107B (2019) 1420–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [370].Adams ME, Atkinson MH, Lussier AJ, Schulz JI, Siminovitch KA, Wade JP, Zummer M, Osteoarthr. Cartil. 3 (1995) 213–225. [DOI] [PubMed] [Google Scholar]
- [371].Rutjes AWS, Jüni P, da Costa BR, Trelle S, Nüesch E, Reichenbach S, Ann. Intern. Med. 157 (2012) 180–191. [DOI] [PubMed] [Google Scholar]
- [372].Higashide T, Sugiyama K, Clin. Ophthalmol. 2 (2008) 21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [373].Harooni M, Freilich JM, Abelson M, Refojo M, Arch. Ophthalmol. 116 (1998) 1218–1221. [DOI] [PubMed] [Google Scholar]
- [374].Hemmrich K, Van de Sijpe K, Rhodes NP, Hunt JA, Di Bartolo C, Pallua N, Blondeel P, von Heimburg D, J. Surg. Res. 144 (2008) 82–88. [DOI] [PubMed] [Google Scholar]
- [375].Turner NJ, Kielty CM, Walker MG, Canfield AE, Biomater. 25 (2004) 5955–5964. [DOI] [PubMed] [Google Scholar]
- [376].Colletta V, Dioguardi D, Di Lonardo A, Maggio G, Torasso F, J. Wound Care 12 (2003) 357–361. [DOI] [PubMed] [Google Scholar]
- [377].Myers SR, Partha VN, Soranzo C, Price RD, Navsaria HA, Tissue Eng. 13 (2007) 2733–2741. [DOI] [PubMed] [Google Scholar]
- [378].ISO 10993, (2018).
- [379].F04.42, ASTM International 13.01 (2020).
- [380].F04.42, ASTM International 13.01 (2019).
- [381].Williams DF, Biomater. 29 (2008) 2941–2953. [DOI] [PubMed] [Google Scholar]
- [382].Williams BR, Gelman RA, Poppke DC, Piez KA, J. Biol. Chem. 253 (1978) 6578–6585. [PubMed] [Google Scholar]
- [383].Rosenblatt J, Devereux B, Wallace D, Biomater. 15 (1994) 985–995. [DOI] [PubMed] [Google Scholar]
- [384].Ramanujan S, Pluen A, McKee TD, Brown EB, Boucher Y, Jain RK, Biophys. J. 83 (2002) 1650–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [385].Erikson A, Andersen HN, Naess SN, Sikorski P, Davies C. de L., Biopolymers 89 (2008) 135–143. [DOI] [PubMed] [Google Scholar]
- [386].Gillette BM, Jensen JA, Wang M, Tchao J, Sia SK, Adv. Mater. 22 (2010) 686–691. [DOI] [PubMed] [Google Scholar]
- [387].Gobeaux F, Mosser G, Anglo A, Panine P, Davidson P, Giraud-Guille M-M, Belamie E, J. Mol. Biol. 376 (2008) 1509–1522. [DOI] [PubMed] [Google Scholar]
- [388].Brightman AO, Rajwa BP, Sturgis JE, McCallister ME, Robinson JP, Voytik-Harbin SL, Biopolymers 54 (2000) 222–234. [DOI] [PubMed] [Google Scholar]
- [389].Bailey JL, Critser PJ, Whittington C, Kuske JL, Yoder MC, Voytik-Harbin SL, Biopolymers 95 (2011) 77–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [390].Miron-Mendoza M, Seemann J, Grinnell F, Biomater. 31 (2010) 6425–6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [391].Fraley SI, Wu P, He L, Feng Y, Krisnamurthy R, Longmore GD, Wirtz D, Sci. Rep. 5 (2015) 14580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [392].Antoine EE, Vlachos PP, Rylander MN, Tissue Eng Part B Rev 20 (2014) 683–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [393].Raub CB, Unruh J, Suresh V, Krasieva T, Lindmo T, Gratton E, Tromberg BJ, George SC, Biophys. J. 94 (2008) 2361–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [394].Sung KE, Su G, Pehlke C, Trier SM, Eliceiri KW, Keely PJ, Friedl A, Beebe DJ, Biomater. 30 (2009) 4833–4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [395].Yamamura N, Sudo R, Ikeda M, Tanishita K, Tissue Eng. 13 (2007) 1443–1453. [DOI] [PubMed] [Google Scholar]
- [396].Raub CB, Suresh V, Krasieva T, Lyubovitsky J, Mih JD, Putnam AJ, Tromberg BJ, George SC, Biophys. J. 92 (2007) 2212–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [397].Yang Y, Motte S, Kaufman LJ, Biomater. 31 (2010) 5678–5688. [DOI] [PubMed] [Google Scholar]
- [398].Chrobak KM, Potter DR, Tien J, Microvasc. Res. 71 (2006) 185–196. [DOI] [PubMed] [Google Scholar]
- [399].Achilli M, Mantovani D, Polym, 2 (2010) 664–680. [Google Scholar]
- [400].Wood GC, Keech MK, Biochem. J. 75 (1960) 588–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [401].Chen C, Loe F, Blocki A, Peng Y, Raghunath M, Adv. Drug Deliv. Rev. 63 (2011) 277–290. [DOI] [PubMed] [Google Scholar]
- [402].Parkinson J, Kadler KE, Brass A, J. Mol. Biol. 247 (1995) 823–831. [DOI] [PubMed] [Google Scholar]
- [403].Lareu RR, Subramhanya KH, Peng Y, Benny P, Chen C, Wang Z, Rajagopalan R, Raghunath M, FEBS Lett. 581 (2007) 2709–2714. [DOI] [PubMed] [Google Scholar]
- [404].Dewavrin J-Y, Hamzavi N, Shim VPW, Raghunath M, Acta Biomater. 10 (2014) 4351–4359. [DOI] [PubMed] [Google Scholar]
- [405].Dewavrin J-Y, Abdurrahiem M, Blocki A, Musib M, Piazza F, Raghunath M, J. Phys. Chem. B 119 (2015) 4350–4358. [DOI] [PubMed] [Google Scholar]
- [406].Saeidi N, Karmelek KP, Paten JA, Zareian R, DiMasi E, Ruberti JW, Biomaterials 33 (2012) 7366–7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [407].Ranamukhaarachchi SK, Modi RN, Han A, Velez DO, Kumar A, Engler AJ, Fraley SI, Biomater. Sci. 7 (2019) 618–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [408].Oryan A, Kamali A, Moshiri A, Baharvand H, Daemi H, Int. J. Biol. Macromol. 107 (2018) 678–688. [DOI] [PubMed] [Google Scholar]
- [409].Jayakrishnan A, Jameela SR, Biomater. 17 (1996) 471–484. [DOI] [PubMed] [Google Scholar]
- [410].Damink L. H. H. Olde, Dijkstra PJ, Van Luyn MJA, Van Wachem PB, Nieuwenhuis P, Feijen J, J. Mater. Sci. Mater. Med. 6 (1995) 460–472. [Google Scholar]
- [411].Peng YY, Glattauer V, Ramshaw JAM, Int. J. Biomater. 2017 (2017) 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [412].Ma B, Wang X, Wu C, Chang J, Regen. Biomater. 1 (2014) 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [413].Kim S-S, Lim S-H, Cho SW, Gwak S-J, Hong Y-S, Chang BC, Park MH, Song KW, Choi CY, Kim B-S, Exp. Mol. Med. 38 (2006) 273–283. [DOI] [PubMed] [Google Scholar]
- [414].Matsuda S, Iwata H, Se N, Ikada Y, J. Biomed. Mater. Res. 45 (1999) 20–27. [DOI] [PubMed] [Google Scholar]
- [415].Kalbitzer L, Franke K, Möller S, Schnabelrauch M, Pompe T, J. Mater. Chem. B 3 (2015) 8902–8910. [DOI] [PubMed] [Google Scholar]
- [416].Samadian H, Vaez A, Ehterami A, Salehi M, Farzamfar S, Sahrapeyma H, Norouzi P, J. Mater. Sci. Mater. Med. 30 (2019) 107. [DOI] [PubMed] [Google Scholar]
- [417].Chan KLS, Khankhel AH, Thompson RL, Coisman BJ, Wong KHK, Truslow JG, Tien J, J. Biomed. Mater. Res. A. 102A (2014) 3186–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [418].Bloemen M, Van Stappen T, Willot P, Lammertyn J, Koeckelberghs G, Geukens N, Gils A, Verbiest T, PLOS ONE 9 (2014) e109475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [419].Pal P, Dadhich P, Kumar Srivas P, Das B, Maulik D, Dhara S, Biomater. Sci. 5 (2017) 1786–1799. [DOI] [PubMed] [Google Scholar]
- [420].Yoo JS, Kim YJ, Kim SH, Choi SH, Korean J Thorac. Cardiovasc. Surg. 44 (2011) 197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [421].Sung H-W, Liang I-L, Chen C-N, Huang R-N, Liang H-F, J. Biomed. Mater. Res. 55 (2001) 538–546. [DOI] [PubMed] [Google Scholar]
- [422].Zhou X, Wang J, Fang W, Tao Y, Zhao T, Xia K, Liang C, Hua J, Li F, Chen Q, Acta Biomater. 71 (2018) 496–509. [DOI] [PubMed] [Google Scholar]
- [423].Zhang Y, Wang Q-S, Yan K, Qi Y, Wang G-F, Cui Y-L, J. Biomed. Mater. Res. A. 104A (2016) 1863–1870. [DOI] [PubMed] [Google Scholar]
- [424].Výborný K, Vallová J, Kočí Z, Kekulová K, Jiráková K, Jendelová P, Hodan J, Kubinová Š, Sci. Rep. 9 (2019) 10674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [425].Wang Y, Bao J, Wu X, Wu Q, Li Y, Zhou Y, Li L, Bu H, Sci. Rep. 6 (2016) 24779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [426].Avery NC, Bailey AJ, Scand. J. Med. Sci. Sports 15 (2005) 231–240. [DOI] [PubMed] [Google Scholar]
- [427].Brownlee M, Annu. Rev. Med. 46 (1995) 223–234. [DOI] [PubMed] [Google Scholar]
- [428].Roy R, Boskey A, Bonassar LJ, J. Biomed. Mater. Res. A 93 (2010) 843–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [429].Jiang Q, Reddy N, Zhang S, Roscioli N, Yang Y, J. Biomed. Mater. Res. A. 101A (2013) 1237–1247. [DOI] [PubMed] [Google Scholar]
- [430].Cumming MH, Leonard AR, LeCorre-Bordes DS, Hofman K, Int. J. Biol. Macromol. 114 (2018) 874–881. [DOI] [PubMed] [Google Scholar]
- [431].Gyawali D, Nair P, Zhang Y, Tran RT, Zhang C, Samchukov M, Makarov M, Kim HKW, Yang J, Biomater. 31 (2010) 9092–9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [432].Tran RT, Zhang Y, Gyawali D, Yang J, Recent Pat. Biomed. Eng. 2 (2009) 216–227. [Google Scholar]
- [433].Sánchez-Ferrero A, Mata Á, Mateos-Timoneda MA, Rodríguez-Cabello JC, Alonso M, Planell J, Engel E, Biomater. 68 (2015) 42–53. [DOI] [PubMed] [Google Scholar]
- [434].Reddy N, Yang Y, Food Chem. 118 (2010) 702–711. [Google Scholar]
- [435].Wang L, Stegemann JP, Acta Biomater. 7 (2011) 2410–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [436].Browe DC, Mahon OR, Díaz- Payno PJ, Cassidy N, Dudurych I, Dunne A, Buckley CT, Kelly DJ, J. Biomed. Mater. Res. A. 107A (2019) 2222–2234. [DOI] [PubMed] [Google Scholar]
- [437].Skopinska-Wisniewska J, Kuderko J, Bajek A, Maj M, Sionkowska A, Ziegler-Borowska M, Mater. Sci. Eng. C. 60 (2016) 100–108. [DOI] [PubMed] [Google Scholar]
- [438].Kwon YS, Lee SH, Hwang YC, Rosa V, Lee KW, Min KS, Int. Endod. J. 50 (2017) 58–66. [DOI] [PubMed] [Google Scholar]
- [439].Koob TJ, Willis TA, Qiu YS, Hernandez DJ, J. Biomed. Mater. Res. 56 (2001) 40–48. [DOI] [PubMed] [Google Scholar]
- [440].Rioja AY, Muniz-Maisonet M, Koob TJ, Gallant ND, AIMS Bioeng. 4 (2017) 300–317. [Google Scholar]
- [441].Lü X, Zhai W, Zhou Y, Zhou Y, Zhang H, Chang J, J. Mater. Sci. Mater. Med. 21 (2010) 473–480. [DOI] [PubMed] [Google Scholar]
- [442].Heijmen FH, du Pont JS, Middelkoop E, Kreis RW, Hoekstra MJ, Biomater. 18 (1997) 749–754. [DOI] [PubMed] [Google Scholar]
- [443].Lee J, Yeo M, Kim W, Koo Y, Kim GH, Int. J. Biol. Macromol. 110 (2018) 497–503. [DOI] [PubMed] [Google Scholar]
- [444].Koh LB, Islam MM, Mitra D, Noel CW, Merrett K, Odorcic S, Fagerholm P, Jackson WB, Liedberg B, Phopase J, Griffith M, J. Funct. Biomater. 4 (2013) 162–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [445].Zeeman R, Dijkstra PJ, van Wachem PB, van Luyn MJA, Hendriks M, Cahalan PT, Feijen J, J. Biomed. Mater. Res. 51 (2000) 541–548. [DOI] [PubMed] [Google Scholar]
- [446].Tu R, Shen S-H, Lin D, Hata C, Thyagarajan K, Noishiki Y, Quijano RC, J. Biomed. Mater. Res. 28 (1994) 677–684. [DOI] [PubMed] [Google Scholar]
- [447].Singh RK, Seliktar D, Putnam AJ, Biomater. 34 (2013) 9331–9340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [448].Voorhaar L, Hoogenboom R, Chem. Soc. Rev. 45 (2016) 4013–4031. [DOI] [PubMed] [Google Scholar]
- [449].Hu W, Wang Z, Xiao Y, Zhang S, Wang J, Biomater. Sci. 7 (2019) 843–855. [DOI] [PubMed] [Google Scholar]
- [450].Zheng Z, Hu J, Wang H, Huang J, Yu Y, Zhang Q, Cheng Y, ACS Appl. Mater. Interfaces 9 (2017) 24511–24517. [DOI] [PubMed] [Google Scholar]
- [451].Zabow G, Dodd SJ, Koretsky AP, Nat. 520 (2015) 73–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [452].Purcell BP, Lobb D, Charati MB, Dorsey SM, Wade RJ, Zellars KN, Doviak H, Pettaway S, Logdon CB, Shuman JA, Freels PD, Gorman Iii JH, Gorman RC, Spinale FG, Burdick JA, Nat. Mater. 13 (2014) 653–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [453].Chung HJ, Lee Y, Park TG, J. Control. Release. 127 (2008) 22–30. [DOI] [PubMed] [Google Scholar]
- [454].Nakahata M, Takashima Y, Yamaguchi H, Harada A, Nat. Commun. 2 (2011) 511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [455].Holten-Andersen N, Harrington MJ, Birkedal H, Lee BP, Messersmith PB, Lee KYC, Waite JH, PNAS 108 (2011) 2651–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [456].Lu HD, Charati MB, Kim IL, Burdick JA, Biomater. 33 (2012) 2145–2153. [DOI] [PubMed] [Google Scholar]
- [457].Chen G, Jiang M, Chem. Soc. Rev. 40 (2011) 2254–2266. [DOI] [PubMed] [Google Scholar]
- [458].Rodell CB, Kaminski AL, Burdick JA, Biomacromolecules 14 (2013) 4125–4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [459].Rosales AM, Rodell CB, Chen MH, Morrow MG, Anseth KS, Burdick JA, Bioconjugate Chem. 29 (2018) 905–913. [DOI] [PubMed] [Google Scholar]
- [460].Montanari E, D’Arrigo G, Di Meo C, Virga A, Coviello T, Passariello C, Matricardi P, Eur. J. Pharm. Biopharm. 87 (2014) 518–523. [DOI] [PubMed] [Google Scholar]
- [461].Jung Y, Park W, Park H, Lee D-K, Na K, Carbohydr. Polym. 156 (2017) 403–408. [DOI] [PubMed] [Google Scholar]
- [462].Laffleur F, Röggla J, Idrees MA, Griessinger J, J. Pharm. Sci. 103 (2014) 2414–2423. [DOI] [PubMed] [Google Scholar]
- [463].Laffleur F, Schmelzle F, Ganner A, Vanicek S, AAPS PharmSciTech 18 (2017) 2102–2109. [DOI] [PubMed] [Google Scholar]
- [464].Tian H, He Z, Sun C, Yang C, Zhao P, Liu L, Leong KW, Mao H-Q, Liu Z, Chen Y, Adv. Healthcare Mater. 7 (2018) 1800285. [DOI] [PubMed] [Google Scholar]
- [465].Chen W, Zou Y, Meng F, Cheng R, Deng C, Feijen J, Zhong Z, Biomacromolecules 15 (2014) 900–907. [DOI] [PubMed] [Google Scholar]
- [466].Trombino S, Servidio C, Curcio F, Cassano R, Pharmaceutics 11 (2019) 407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [467].Chauvin J-PR, Pratt DA, Angew. Chem. Int. Ed. 56 (2017) 6255–6259. [DOI] [PubMed] [Google Scholar]
- [468].Peng H-H, Chen Y-M, Lee C-I, Lee M-W, J. Mater. Sci. Mater. Med. 24 (2013) 1375–1382. [DOI] [PubMed] [Google Scholar]
- [469].Bermejo-Velasco D, Azémar A, Oommen OP, Hilborn J, Varghese OP, Biomacromolecules 20 (2019) 1412–1420. [DOI] [PubMed] [Google Scholar]
- [470].Williams DL, Mann BK, Int. J. Biomater. 2013 (2013) 460437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [471].Kafedjiiski K, Jetti RKR, Foger F, Herbert H, Werle M, Hoffer M, Bernkop-Schnurch A ¨, Int. J. Pharm. 343 (2007) 48–58. [DOI] [PubMed] [Google Scholar]
- [472].Griffith GL, Wirostko B, Lee H-K, Cornell LE, McDaniel JS, Zamora DO, Johnson AJ, Burns 44 (2018) 1179–1186. [DOI] [PubMed] [Google Scholar]
- [473].Yang G, Prestwich GD, Mann BK, ISRN Vet. Sci. 2011 (2011) e851593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [474].Liu Y, Shu XZ, Prestwich GD, Tissue Eng. 12 (2006) 3405–3416. [DOI] [PubMed] [Google Scholar]
- [475].Bian S, He M, Sui J, Cai H, Sun Y, Liang J, Fan Y, Zhang X, Colloids Surf. B: Biointerfaces 140 (2016) 392–402. [DOI] [PubMed] [Google Scholar]
- [476].Hozumi T, Kageyama T, Ohta S, Fukuda J, Ito T, Biomacromolecules 19 (2018) 288–297. [DOI] [PubMed] [Google Scholar]
- [477].Xu J, Liu Y, Hsu S, Molecules 24 (2019) 3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [478].Hassib HB, Abdel-Kader NS, Issa YM, J. Solution Chem. 41 (2012) 2036–2046. [Google Scholar]
- [479].Khunmanee S, Jeong Y, Park H, J. Tissue Eng. 8 (2017) doi: 10.1177/2041731417726464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [480].Ma X, Xu T, Chen W, Qin H, Chi B, Ye Z, Carbohydr. Polym. 179 (2018) 100–109. [DOI] [PubMed] [Google Scholar]
- [481].Bajaj G, Kim MR, Mohammed SI, Yeo Y, Control J. Release 158 (2012) 386–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [482].Wang H, Zhu D, Paul A, Cai L, Enejder A, Yang F, Heilshorn SC, Adv. Funct. Mater. 27 (2017) 1605609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [483].Ifkovits JL, Burdick JA, Tissue Eng. 13 (2007) 2369–2385. [DOI] [PubMed] [Google Scholar]
- [484].Ibrahim S, Kothapalli CR, Kang QK, Ramamurthi A, Acta Biomater. 7 (2011) 653–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [485].Tous E, Ifkovits JL, Koomalsingh KJ, Shuto T, Soeda T, Kondo N, Gorman JH, Gorman RC, Burdick JA, Biomacromolecules 12 (2011) 4127–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [486].Poldervaart MT, Goversen B, de Ruijter M, Abbadessa A, Melchels FPW, Öner FC, Dhert WJA, Vermonden T, Alblas J, PLoS One 12 (2017) e0177628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [487].Park H, Choi B, Hu J, Lee M, Acta Biomater. 9 (2013) 4779–4786. [DOI] [PubMed] [Google Scholar]
- [488].Fenn SL, Oldinski RA, J. Biomed. Mater. Res. Part B Appl. Biomater. 104 (2016) 1229–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [489].Bencherif SA, Srinivasan A, Horkay F, Hollinger JO, Matyjaszewski K, Washburn NR, Biomaterials 29 (2008) 1739–1749. [DOI] [PubMed] [Google Scholar]
- [490].Ananthanarayanan B, Kim Y, Kumar S, Biomaterials 32 (2011) 7913–7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [491].Gwon K, Kim E, Tae G, Acta Biomater. 49 (2017) 284–295. [DOI] [PubMed] [Google Scholar]
- [492].Das D, Pham TTH, Noh I, Colloids Surf. B 170 (2018) 64–75. [DOI] [PubMed] [Google Scholar]
- [493].Larrañeta E, Henry M, Irwin NJ, Trotter J, Perminova AA, Donnelly RF, Carbohydr. Polym. 181 (2018) 1194–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [494].Lee F, Lim J, Reithofer MR, Seong Lee S, Eun Chung J, Hauser CAE, Kurisawa M, Polym. Chem. 6 (2015) 4462–4472. [Google Scholar]
- [495].Nakama T, Ooya T, Yui N, Polym. J. 36 (2004) 338–344. [Google Scholar]
- [496].Malson T, Lindqvist BL, Gel of Crosslinked Hyaluronic Acid for Use as A Vitreous Humor Substitute, 4,716,154, n.d. [Google Scholar]
- [497].Xue Y, Chen H, Xu C, Yu D, Xu H, Hu Y, RSC Adv. 10 (2020) 7206–7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [498].Borzacchiello A, Luisa R, Malle BM, Schwach-Abdellaoui K, Ambrosio L, BioMed Res. Int. 2015 (2015) 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [499].Longin F, Schwach-Abdellaoui K., Method of Cross-Linking Hyaluroncacid with Divinylsulfone, 2013/0338100 A1, 2013. [Google Scholar]
- [500].Kolb HC, Finn MG, Sharpless KB, Angew. Chem. Int. Ed. 40 (2001) 2004–2021. [DOI] [PubMed] [Google Scholar]
- [501].Crescenzi V, Cornelio L, Di Meo C, Nardecchia S, Lamanna R, Biomacromolecules 8 (2007) 1844–1850. [DOI] [PubMed] [Google Scholar]
- [502].Nimmo CM, Owen SC, Shoichet MS, Biomacromolecules 12 (2011) 824–830. [DOI] [PubMed] [Google Scholar]
- [503].Tan H, Rubin JP, Marra KG, Macromol. Rapid Commun. 32 (2011) 905–911. [DOI] [PubMed] [Google Scholar]
- [504].Yu F, Cao X, Li Y, Zeng L, Zhu J, Wang G, Chen X, Polym. Chem. 5 (2014) 5116–5123. [Google Scholar]
- [505].Xing D, Ma L, Gao C, J. Bioact. Compat. Polym. 32 (2017) 382–396. [Google Scholar]
- [506].Smith LJ, Taimoory SM, Tam RY, Baker AEG, Binth Mohammad N, Trant JF, Shoichet MS, Biomacromolecules 19 (2018) 926–935. [DOI] [PubMed] [Google Scholar]
- [507].Yu F, Cao X, Li Y, Zeng L, Yuan B, Chen X, Polym. Chem. 5 (2013) 1082–1090. [Google Scholar]
- [508].Wang G, Cao X, Dong H, Zeng L, Yu C, Chen X, Polymers 10 (2018) 949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [509].Wu H, Devaraj NK, Top. Curr. Chem. 374 (2016) 3. [DOI] [PubMed] [Google Scholar]
- [510].Delplace V, Nickerson PEB, Ortin- Martinez A, Baker AEG, Wallace VA, Shoichet MS, Adv. Funct. Mater. 30 (2020) 1903978. [Google Scholar]
- [511].Yu F, Cao X, Li Y, Chen X, ACS Macro Lett. 4 (2015) 289–292. [DOI] [PubMed] [Google Scholar]
- [512].Rostovtsev VV, Green LG, Fokin VV, Sharpless KB, Angew. Chem. Int. Ed. 41 (2002) 2596–2599. [DOI] [PubMed] [Google Scholar]
- [513].Huerta-Angeles G, Šmejkalová D, Chládková D, Ehlová T, Buffa R, Velebný V, Carbohydr. Polym. 84 (2011) 1293–1300. [Google Scholar]
- [514].Fu S, Dong H, Deng X, Zhuo R, Zhong Z, Carbohydr. Polym. 169 (2017) 332–340. [DOI] [PubMed] [Google Scholar]
- [515].Takahashi A, Suzuki Y, Suhara T, Omichi K, Shimizu A, Hasegawa K, Kokudo N, Ohta S, Ito T, Biomacromolecules 14 (2013) 3581–3588. [DOI] [PubMed] [Google Scholar]
- [516].Han S-S, Yoon HY, Yhee JY, Cho MO, Shim H-E, Jeong J-E, Lee D-E, Kim K, Guim H, Lee JH, Huh KM, Kang S-W, Polym. Chem. 9 (2017) 20–27. [Google Scholar]
- [517].Kurisawa M, Lee F, Wang L-S, Chung JE, J. Mater. Chem. 20 (2010) 5371. [Google Scholar]
- [518].Lee F, Chung JE, Kurisawa M, Control J. Release 134 (2009) 186–193. [DOI] [PubMed] [Google Scholar]
- [519].Jin R, Moreira Teixeira LS, Dijkstra PJ, van Blitterswijk CA, Karperien M, Feijen J, Biomaterials 31 (2010) 3103–3113. [DOI] [PubMed] [Google Scholar]
- [520].Wennink JWH, Niederer K, Bochyńska AI, Moreira Teixeira LS, Karperien M, Feijen J, Dijkstra PJ, Macromol. Symp. 309–310 (2011) 213–221. [Google Scholar]
- [521].Wang L-S, Chung JE, Pui-Yik Chan P, Kurisawa M, Biomaterials 31 (2010) 1148–1157. [DOI] [PubMed] [Google Scholar]
- [522].Kurisawa M, Chung JE, Yang YY, Gao SJ, Uyama H, Chem. Commun. (2005) 4312–4314. [DOI] [PubMed] [Google Scholar]
- [523].Shona Pek Y, Kurisawa M, Gao S, Chung JE, Ying JY, Biomaterials 30 (2009) 822–828. [DOI] [PubMed] [Google Scholar]
- [524].Xu K, Narayanan K, Lee F, Bae KH, Gao S, Kurisawa M, Acta Biomater. 24 (2015) 159–171. [DOI] [PubMed] [Google Scholar]
- [525].Roberts JJ, Naudiyal P, Lim KS, Poole-Warren LA, Martens PJ, Biomater. Res. 20 (2016) 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [526].Raia NR, Partlow BP, McGill M, Kimmerling EP, Ghezzi CE, Kaplan DL, Biomaterials 131 (2017) 58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [527].Fan Z, Zhang Y, Fang S, Xu C, Li X, RSC Adv. 5 (2015) 1929–1936. [Google Scholar]
- [528].Ganesh N, Hanna C, Nair SV, Nair LS, Int. J. Biol. Macromol. 55 (2013) 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [529].Spadiut O, Herwig C, Pharm. Bioprocess. 1 (2013) 283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [530].Sakai S, Moriyama K, Taguchi K, Kawakami K, Biomacromolecules 11 (2010) 2179–2183. [DOI] [PubMed] [Google Scholar]
- [531].Akkara JA, Wang J, Yang D-P, Gonsalves KE, Macromolecules 33 (2000) 2377–2382. [Google Scholar]
- [532].Ryu JH, Lee Y, Do MJ, Jo SD, Kim JS, Kim B-S, Im G-I, Park TG, Lee H, Acta Biomater. 10 (2014) 224–233. [DOI] [PubMed] [Google Scholar]
- [533].Wang H, Heilshorn SC, Adv. Mater. 27 (2015) 3717–3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [534].Rosales AM, Anseth KS, Nat. Rev. Mater. 1 (2016) 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [535].Vázquez-Portalatín N, Kilmer CE, Panitch A, Liu JC, Biomacromolecules 17 (2016) 3145–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [536].Tsaryk R, Gloria A, Russo T, Anspach L, De Santis R, Ghanaati S, Unger RE, Ambrosio L, Kirkpatrick CJ, Acta Biomater. 20 (2015) 10–21. [DOI] [PubMed] [Google Scholar]
- [537].Xin X, Borzacchiello A, Netti PA, Ambrosio L, Nicolais L, J. Biomater. Sci. Polym. Ed. 15 (2004) 1223–1236. [DOI] [PubMed] [Google Scholar]
- [538].Avendano A, Chang JJ, Cortes-Medina MG, Seibel AJ, Admasu BR, Boutelle CM, Bushman AR, Garg AA, DeShetler CM, Cole SL, Song JW, ACS Biomater. Sci. Eng. 6 (2020) 1408–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [539].Lai VK, Nedrelow DS, Lake SP, Kim B, Weiss EM, Tranquillo RT, Barocas VH, Ann. Biomed. Eng. 44 (2016) 2984–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [540].Federico S, Nöchel U, Löwenberg C, Lendlein A, Neffe AT, Acta Biomater. 38 (2016) 1–10. [DOI] [PubMed] [Google Scholar]
- [541].Kontturi L-S, Järvinen E, Muhonen V, Collin EC, Pandit AS, Kiviranta I, Yliperttula M, Urtti A, Drug Deliv. and Transl. Res. 4 (2014) 149–158. [DOI] [PubMed] [Google Scholar]
- [542].Saraswathy K, Agarwal G, Srivastava A, J. Appl. Polym. Sci. 137 (2020) e49285. [Google Scholar]
- [543].Heo J, Koh RH, Shim W, Kim HD, Yim H-G, Hwang NS, Drug Deliv. Transl. Res. 6 (2016) 148–158. [DOI] [PubMed] [Google Scholar]
- [544].Perry CC, in: Meyers RA (Ed.), Encyclopedia of Physical Science and Technology (Third Edition), Academic Press, New York, 2003, pp. 173–191. [Google Scholar]
- [545].Kadler KE, Hojima Y, Prockop DJ, J. Biol. Chem. 262 (1987) 15696–15701. [PubMed] [Google Scholar]
- [546].Zhu M, Feng Q, Sun Y, Li G, Bian L, J. Biomed. Mater. Res. 105 (2017) 2292–2300. [DOI] [PubMed] [Google Scholar]
- [547].Jeong GS, Kwon GH, Kang AR, Jung BY, Park Y, Chung S, Lee S-H, Biomed. Microdevices 13 (2011) 717–723. [DOI] [PubMed] [Google Scholar]
- [548].Lou J, Stowers R, Nam S, Xia Y, Chaudhuri O, Biomaterials 154 (2018) 213–222. [DOI] [PubMed] [Google Scholar]
- [549].Kim J-K, Lee J-S, Jung H-J, Cho J-H, Heo J-I, Chang Y-H, J. Nanosci. Nanotechnol. 7 (2007) 3852–3856. [DOI] [PubMed] [Google Scholar]
- [550].Kim HJ, Kim KK, Park IK, Choi BS, Kim JH, Kim MS, Tissue Eng. Regen. Med. 9 (2012) 57–62. [Google Scholar]
- [551].Calderon L, Collin E, Velasco-Bayon D, Murphy M, O’Halloran D, Pandit A, Eur. Cells Mater. 20 (2010) 134–148. [DOI] [PubMed] [Google Scholar]
- [552].He Z, Xiong L, J. Macromol. Sci. B 52 (2013) 1626–1635. [Google Scholar]
- [553].Priyadarshani P, Li Y, Yang S, Yao L, J. Biomed. Mater. Res. Part A 104 (2016) 419–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [554].Mohammadi F, Mohammadi Samani S, Tanideh N, Ahmadi F, Adv. Pharm. Bull. 8 (2018) 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [555].Ying H, Zhou J, Wang M, Su D, Ma Q, Lv G, Chen J, Mater. Sci. Eng. C 101 (2019) 487–498. [DOI] [PubMed] [Google Scholar]
- [556].Zhang T, Chen H, Zhang Y, Zan Y, Ni T, Liu M, Pei R, Colloids Surf. B 174 (2019) 528–535. [DOI] [PubMed] [Google Scholar]
- [557].Eng D, Caplan M, Preul M, Panitch A, Acta Biomaterialia 6 (2010) 2407–2414. [DOI] [PubMed] [Google Scholar]
- [558].Suri S, Schmidt CE, Tissue Eng. Part A 16 (2010) 1703–1716. [DOI] [PubMed] [Google Scholar]
- [559].Geissler SA, Sabin AL, Besser RR, Gooden OM, Shirk BD, Nguyen QM, Khaing ZZ, Schmidt CE, J. Neural Eng. 15 (2018) 025004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [560].Spearman BS, Agrawal NK, Rubiano A, Simmons CS, Mobini S, Schmidt CE, J. Biomed. Mater. Res. Part A 108 (2020) 279–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [561].Guo Y, Yuan T, Xiao Z, Tang P, Xiao Y, Fan Y, Zhang X, J. Mater. Sci. Mater. Med. 23 (2012) 2267–2279. [DOI] [PubMed] [Google Scholar]
- [562].Zhang L, Li K, Xiao W, Zheng L, Xiao Y, Fan H, Zhang X, Carbohydr. Polym. 84 (2011) 118–125. [Google Scholar]
- [563].Gao Y, Li K, Guo L, Fan H, Fan Y, Zhang X, Mater. Lett. 258 (2020) 126660. [Google Scholar]
- [564].Kenar H, Ozdogan CY, Dumlu C, Doger E, Kose GT, Hasirci V, Mater. Sci. Eng. C 97 (2019) 31–44. [DOI] [PubMed] [Google Scholar]
- [565].Wu X, Li W, Chen K, Zhang D, Xu L, Yang X, Mater. Today Commun. 21 (2019) 100702. [Google Scholar]
- [566].Xie J, Fan D, Int. J. Polym. Mater. Polym. Biomat. 69 (2020) 928–937. [Google Scholar]
- [567].Gilarska A, Lewandowska-Łańcucka J, Horak W, Nowakowska M, Colloids Surf. B 170 (2018) 152–162. [DOI] [PubMed] [Google Scholar]
- [568].Lewandowska-Łańcucka J, Gilarska A, Buła A, Horak W, Łatkiewicz A, Nowakowska M, Int. J. Biol. Macromol. 136 (2019) 1196–1208. [DOI] [PubMed] [Google Scholar]
- [569].Mahapatra C, Jin G-Z, Kim H-W, Tissue Eng. Regen. Med. 13 (2016) 538–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [570].Fratzl P, Misof K, Zizak I, Rapp G, Amenitsch H, Bernstorff S, J. Struct. Biol. 122 (1998) 119–122. [DOI] [PubMed] [Google Scholar]
- [571].Currey J, Osteoporosis Int. 14 (2003) 29–36. [Google Scholar]
- [572].Provenzano PP, Inman DR, Eliceiri KW, Knittel JG, Yan L, Rueden CT, White JG, Keely PJ, BMC Med. 6 (2008) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [573].Drury JL, Mooney DJ, Biomaterials 24 (2003) 4337–4351. [DOI] [PubMed] [Google Scholar]
- [574].Antoine EE, Vlachos PP, Rylander MN, PLoS One 10 (2015) e0122500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [575].Christiansen DL, Huang EK, Silver FH, Matrix Biol. 19 (2000) 409–420. [DOI] [PubMed] [Google Scholar]
- [576].Park S-N, Park J-C, Kim HO, Song MJ, Suh H, Biomaterials 23 (2002) 1205–1212. [DOI] [PubMed] [Google Scholar]
- [577].Fratzl P, Weinkamer R, Prog. Mater. Sci 52 (2007) 1263–1334. [Google Scholar]
- [578].Cowman MK, Schmidt TA, Raghavan P, Stecco A, F1000Res. 4 (2015) 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [579].Xu B, Li H, Zhang Y, Biomatter 3 (2013) e24651–1-e24651–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [580].Huang D, Huang Y, Xiao Y, Yang X, Lin H, Feng G, Zhu X, Zhang X, Acta Biomater. 97 (2019) 74–92. [DOI] [PubMed] [Google Scholar]
- [581].Silver FH, Freeman JW, Horvath I, Landis WJ, Biomacromolecules 2 (2001) 750–756. [DOI] [PubMed] [Google Scholar]
- [582].Alexander RM, Am. Zool. 24 (1984) 85–94. [Google Scholar]
- [583].Silver FH, Landis WJ, in: Fratzl P (Ed.), Collagen: Structure and Mechanics, Springer US, Boston, MA, 2008, pp. 133–154. [Google Scholar]
- [584].Shen ZL, Kahn H, Ballarini R, Eppell SJ, Biophys. J. 100 (2011) 3008–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [585].Svensson RB, Hassenkam T, Hansen P, Peter Magnusson S, J. Mech. Behav. Biomed. Mater. 3 (2010) 112–115. [DOI] [PubMed] [Google Scholar]
- [586].Kim D-G, Huja SS, Lee HR, Tee BC, Hueni S, J. Biomech. Eng. 132 (2010) 024502. [DOI] [PubMed] [Google Scholar]
- [587].Silver FH, Ebrahimi A, Snowhill PB, Connect. Tissue Res. 43 (2002) 569–580. [PubMed] [Google Scholar]
- [588].Scott JE, Biochem. J. 252 (1988) 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [589].Chen S, Hirota N, Okuda M, Takeguchi M, Kobayashi H, Hanagata N, Ikoma T, Acta Biomater. 7 (2011) 644–652. [DOI] [PubMed] [Google Scholar]
- [590].Holmes DF, Lu Y, Starborg T, Kadler KE, in: Current Topics in Developmental Biology, Elsevier, 2018, pp. 107–142. [DOI] [PubMed] [Google Scholar]
- [591].Garvin KA, VanderBurgh J, Hocking DC, Dalecki D, J. Acoust. Soc. Am. 134 (2013) 1491–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [592].Parry DAD, Biophys. Chem. 29 (1988) 195–209. [DOI] [PubMed] [Google Scholar]
- [593].Roeder BA, Kokini K, Sturgis JE, Robinson JP, Voytik-Harbin SL, J. Biomech. Eng. 124 (2002) 214–222. [DOI] [PubMed] [Google Scholar]
- [594].Folkhard W, Mosler E, Geercken W, Knrzer E, Nemetschek-Gansler H, Nemetschek T, Koch MHJ, Int. J. Biol. Macromol. 9 (1987) 169–175. [Google Scholar]
- [595].Silver FH, Freeman JW, Seehra GP, J. Biomech. 36 (2003) 1529–1553. [DOI] [PubMed] [Google Scholar]
- [596].Hulmes DJS, in: Fratzl P (Ed.), Collagen: Structure and Mechanics, Springer US, Boston, MA, 2008, pp. 15–47. [Google Scholar]
- [597].Avery NC, Bailey AJ, in: Fratzl P (Ed.), Collagen: Structure and Mechanics, Springer US, Boston, MA, 2008, pp. 81–110. [Google Scholar]
- [598].Richardson SH, Starborg T, Lu Y, Humphries SM, Meadows RS, Kadler KE, Mol. Cell. Biol. 27 (2007) 6218–6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [599].Ng CP, Hinz B, Swartz MA, J. Cell Sci. 118 (2005) 4731–4739. [DOI] [PubMed] [Google Scholar]
- [600].Antman-Passig M, Levy S, Gartenberg C, Schori H, Shefi O, Tissue Eng. Part A 23 (2017) 403–414. [DOI] [PubMed] [Google Scholar]
- [601].Vader D, Kabla A, Weitz D, Mahadevan L, PLoS One 4 (2009) e5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [602].Taufalele PV, VanderBurgh JA, Muñoz A, Zanotelli MR, Reinhart-King CA, PLoS One 14 (2019) e0216537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [603].Lanfer B, Freudenberg U, Zimmermann R, Stamov D, Körber V, Werner C, Biomaterials 29 (2008) 3888–3895. [DOI] [PubMed] [Google Scholar]
- [604].Julias M, Edgar LT, Buettner HM, Shreiber DI, BioMed Eng. Online 7 (2008) 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [605].Julias M, Buettner HM, Shreiber DI, Anat. 294 (2011) 243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [606].Pelham RJ, Wang Y, Proc. Natl. Acad. Sci. U. S. A. 94 (1997) 13661–13665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [607].Discher DE, Science 310 (2005) 1139–1143. [DOI] [PubMed] [Google Scholar]
- [608].Junqueira LCU, Toledo OMS, Montes GS, Cell Tissue Res. 217 (1981) 171–175. [DOI] [PubMed] [Google Scholar]
- [609].Vesentini S, Redaelli A, Montevecchi FM, J. Biomech. 38 (2005) 433–443. [DOI] [PubMed] [Google Scholar]
- [610].Meek KM, in: Fratzl P (Ed.), Collagen: Structure and Mechanics, Springer US, Boston, MA, 2008, pp. 359–396. [Google Scholar]
- [611].Wess TJ, in: Fratzl P (Ed.), Collagen: Structure and Mechanics, Springer US, Boston, MA, 2008, pp. 49–80. [Google Scholar]
- [612].Entekhabi E, Haghbin Nazarpak M, Shafieian M, Mohammadi H, Firouzi M, Hassannejad Z, J. Biomed. Mater. Res. Part A (2020) jbm.a.37023. [DOI] [PubMed] [Google Scholar]
- [613].Yang Y, Sun C, Wilhelm ME, Fox LJ, Zhu J, Kaufman LJ, Biomaterials 32 (2011) 7932–7940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [614].Stuart K, Panitch A, Biopolymers 89 (2008) 841–851. [DOI] [PubMed] [Google Scholar]
- [615].Kreger ST, Voytik-Harbin SL, Matrix Biol. 28 (2009) 336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [616].Chen Y, Sui J, Wang Q, Yin Y, Liu J, Wang Q, Han X, Sun Y, Fan Y, Zhang X, Carbohydr. Polym. 190 (2018) 57–66. [DOI] [PubMed] [Google Scholar]
- [617].Wang T-W, Spector M, Acta Biomater. 5 (2009) 2371–2384. [DOI] [PubMed] [Google Scholar]
- [618].Gurkan UA, Tasoglu S, Kavaz D, Demirel MC, Demirci U, Adv. Healthc. Mater. 1 (2012) 149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [619].Thomas D, Fontana G, Chen X, Sanz-Nogués C, Zeugolis DI, Dockery P, O’Brien T, Pandit A, Biomaterials 35 (2014) 8757–8766. [DOI] [PubMed] [Google Scholar]
- [620].Yeh J, Ling Y, Karp JM, Gantz J, Chandawarkar A, Eng G, Blumling III J, Langer R, Khademhosseini A, Biomaterials 27 (2006) 5391–5398. [DOI] [PubMed] [Google Scholar]
- [621].Kupper S, Kłosowska-Chomiczewska I, Szumała P, Carbohydr. Polym. 175 (2017) 347–354. [DOI] [PubMed] [Google Scholar]
- [622].Lim HJ, Cho EC, Lee JA, Kim J, Colloids Surf. A Physicochem Eng. Asp. 402 (2012) 80–87. [Google Scholar]
- [623].Ilgin P, Avci G, Silan C, Ekici S, Aktas N, Ayyala RS, John VT, Sahiner N, Carbohydr. Polym. 82 (2010) 997–1003. [Google Scholar]
- [624].Jia X, Yeo Y, Clifton RJ, Jiao T, Kohane DS, Kobler JB, Zeitels SM, Langer R, Biomacromolecules 7 (2006) 3336–3344. [DOI] [PubMed] [Google Scholar]
- [625].Kamperman T, Henke S, van den Berg A, Shin SR, Tamayol A, Khademhosseini A, Karperien M, Leijten J, Adv. Healthc. Mater. 6 (2017) 1600913. [DOI] [PubMed] [Google Scholar]
- [626].Ma T, Gao X, Dong H, He H, Cao X, Appl. Mater. Today 9 (2017) 49–59. [Google Scholar]
- [627].Sideris E, Griffin DR, Ding Y, Li S, Weaver WM, Di Carlo D, Hsiai T, Segura T, ACS Biomater. Sci. Eng. 2 (2016) 2034–2041. [DOI] [PubMed] [Google Scholar]
- [628].Fontana G, Thomas D, Collin E, Pandit A, Adv. Healthc. Mater. 3 (2014) 2012–2022. [DOI] [PubMed] [Google Scholar]
- [629].Highley CB, Song KH, Daly AC, Burdick JA, Adv. Healthc. Mater. (2020) 1801076. [Google Scholar]
- [630].Murphy SV, Atala A, Nat. Biotechnol. 32 (2014) 773–785. [DOI] [PubMed] [Google Scholar]
- [631].Gillispie G, Prim P, Copus J, Fisher J, Mikos AG, Yoo JJ, Atala A, Lee SJ, Biofabrication 12 (2020) 022003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [632].Derakhshanfar S, Mbeleck R, Xu K, Zhang X, Zhong W, Xing M, Bioact. Mater. 3 (2018) 144–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [633].Yu C, Schimelman J, Wang P, Miller KL, Ma X, You S, Guan J, Sun B, Zhu W, Chen S, Chem. Rev. (2020) 10695–10743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [634].Kim M, Kim GH, Chem. Eng. J. 279 (2015) 317–326. [Google Scholar]
- [635].Zhao X, He J, Xu F, Liu Y, Li D, Virtual Phys. Prototyp. 11 (2016) 57–63. [Google Scholar]
- [636].Negro A, Cherbuin T, Lutolf MP, Sci. Rep. 8 (2018) 17099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [637].Li J, Chen M, Fan X, Zhou H, J. Transl. Med. 14 (2016) 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [638].Nocera AD, Comín R, Salvatierra NA, Cid MP, Biomed. Microdevices 20 (2018) 26. [DOI] [PubMed] [Google Scholar]
- [639].Moncal KK, Ozbolat V, Datta P, Heo DN, Ozbolat IT, J. Mater. Sci. Mater. Med. 30 (2019) 55. [DOI] [PubMed] [Google Scholar]
- [640].Ouyang L, Highley CB, Rodell CB, Sun W, Burdick JA, ACS Biomater. Sci. Eng. 2 (2016) 1743–1751. [DOI] [PubMed] [Google Scholar]
- [641].Lam T, Dehne T, Krüger JP, Hondke S, Endres M, Thomas A, Lauster R, Sittinger M, Kloke L, J. Biomed. Mater. Res. Part B Appl. Biomater. 107 (2019) 2649–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [642].Mazzocchi A, Devarasetty M, Huntwork R, Soker S, Skardal A, Biofabrication 11 (2018) 015003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [643].Fischer RL, McCoy MG, Grant SA, J. Mater. Sci. Mater. Med. 23 (2012) 1645–1654. [DOI] [PubMed] [Google Scholar]
- [644].Zhang K, Fan Y, Dunne N, Li X, Regen. Biomater. 5 (2018) 115–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [645].Kim TG, Park TG, Biotechnol. Prog. 22 (2006) 1108–1113. [DOI] [PubMed] [Google Scholar]
- [646].Davidenko N, Campbell JJ, Thian ES, Watson CJ, Cameron RE, Acta Biomater. 6 (2010) 3957–3968. [DOI] [PubMed] [Google Scholar]
- [647].Brown RA, Exp. Cell Res. 319 (2013) 2460–2469. [DOI] [PubMed] [Google Scholar]
- [648].Dash BC, Setia O, Gorecka J, Peyvandi H, Duan K, Lopes L, Nie J, Berthiaume F, Dardik A, Hsia HC, Cells 9 (2020) 966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [649].Hong H, Park SM, Kim D, Park SJ, Kim DS, J. Biomed. Mater. Res. Part B Appl. Biomater. 108 (2020) 1000–1009. [DOI] [PubMed] [Google Scholar]
- [650].Witt J, Borrelli M, Mertsch S, Geerling G, Spaniol K, Schrader S, Tissue Eng. Part A 25 (2018) 1084–1095. [DOI] [PubMed] [Google Scholar]
- [651].Andriakopoulou CE, Zadpoor AA, Grant MH, Riches PE, Mater. Sci. Eng. C 84 (2018) 243–247. [DOI] [PubMed] [Google Scholar]
- [652].Chamieh F, Collignon A-M, Coyac BR, Lesieur J, Ribes S, Sadoine J, Llorens A, Nicoletti A, Letourneur D, Colombier M-L, Nazhat SN, Bouchard P, Chaussain C, Rochefort GY, Sci. Rep. 6 (2016) 38814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [653].Rice JJ, Martino MM, Laporte LD, Tortelli F, Briquez PS, Hubbell JA, Adv. Healthc. Mater. 2 (2013) 57–71. [DOI] [PubMed] [Google Scholar]
- [654].DeForest CA, Polizzotti BD, Anseth KS, Nat. Mater. 8 (2009) 659–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [655].Godesky MD, Shreiber DI, Biointerphases 14 (2019) 061005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [656].Daley WP, Peters SB, Larsen M, J. Cell Sci. 121 (2008) 255–264. [DOI] [PubMed] [Google Scholar]
- [657].Burdick JA, Murphy WL, Nat. Commun. 3 (2012) 1269. [DOI] [PubMed] [Google Scholar]
- [658].Guvendiren M, Burdick JA, Nat. Commun. 3 (2012) 792. [DOI] [PubMed] [Google Scholar]
- [659].Leijten J, Seo J, Yue K, Trujillo-de Santiago G, Tamayol A, Ruiz-Esparza GU, Shin SR, Sharifi R, Noshadi I, Álvarez MM, Zhang YS, Khademhosseini A, Mater. Sci. Eng. R-Rep. 119 (2017) 1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [660].Patterson J, Siew R, Herring SW, Lin ASP, Guldberg R, Stayton PS, Biomaterials 31 (2010) 6772–6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [661].Fisher SA, Anandakumaran PN, Owen SC, Shoichet MS, Adv. Funct. Mater. 25 (2015) 7163–7172. [Google Scholar]
- [662].Oh EJ, Kang S-W, Kim B-S, Jiang G, Cho IH, Hahn SK, J. Biomed. Mater. Res. Part A 86A (2008) 685–693. [DOI] [PubMed] [Google Scholar]
- [663].Kim IL, Mauck RL, Burdick JA, Biomaterials 32 (2011) 8771–8782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [664].Kharkar PM, Kiick KL, Kloxin AM, Chem. Soc. Rev. 42 (2013) 7335–7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [665].Goissis G, Marcantonio E, Marcantônio RAC, Lia RCC, Cancian DCJ, de Carvalho WM, Biomaterials 20 (1999) 27–34. [DOI] [PubMed] [Google Scholar]
- [666].Olde Damink LH, Dijkstra PJ, van Luyn MJ, van Wachem PB, Nieuwenhuis P, Feijen J, Biomaterials 17 (1996) 679–684. [DOI] [PubMed] [Google Scholar]
- [667].Sahoo S, Chung C, Khetan S, Burdick JA, Biomacromolecules 9 (2008) 1088–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [668].Dollinger BR, Gupta MK, Martin JR, Duvall Craig.L., Tissue Eng. Part A 23 (2017) 1120–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [669].Parmar PA, Skaalure SC, Chow LW, St-Pierre J-P, Stoichevska V, Peng YY, Werkmeister JA, Ramshaw JAM, Stevens MM, Biomaterials 99 (2016) 56–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [670].Khetan S, Katz JS, Burdick JA, Soft Matter 5 (2009) 1601. [Google Scholar]
- [671].Fu C, Li H, Li N, Miao X, Xie M, Du W, Zhang L-M, Carbohydr. Polym. 128 (2015) 163–170. [DOI] [PubMed] [Google Scholar]
- [672].Cao W, Li X, Zuo X, Gao C, Regen Biomater 6 (2019) 259–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [673].Mørch ÝA, Donati I, Strand BL, Skjåk-Bræk G, Biomacromolecules 7 (2006) 1471–1480. [DOI] [PubMed] [Google Scholar]
- [674].Suri S, Schmidt CE, Acta Biomater. 5 (2009) 2385–2397. [DOI] [PubMed] [Google Scholar]
- [675].Khetan S, Burdick JA, Biomaterials 31 (2010) 8228–8234. [DOI] [PubMed] [Google Scholar]
- [676].Gramlich WM, Kim IL, Burdick JA, Biomaterials 34 (2013) 9803–9811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [677].Marklein RA, Burdick JA, Soft Matter 6 (2009) 136–143. [Google Scholar]
- [678].Mohamed MA, Fallahi A, El-Sokkary AMA, Salehi S, Akl MA, Jafari A, Tamayol A, Fenniri H, Khademhosseini A, Andreadis ST, Cheng C, Prog. Polym. Sci. 98 (2019) 101147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [679].Ferreira AM, Gentile P, Chiono V, Ciardelli G, Acta Biomater. 8 (2012) 3191–3200. [DOI] [PubMed] [Google Scholar]
- [680].Wang L, Stegemann JP, Biomaterials 31 (2010) 3976–3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [681].Reichert JC, Heymer A, Berner A, Eulert J, Nöth U, Biomed. Mater. 4 (2009) 065001. [DOI] [PubMed] [Google Scholar]
- [682].Haberstroh K, Ritter K, Kuschnierz J, Bormann K-H, Kaps C, Carvalho C, Mülhaupt R, Sittinger M, Gellrich N-C, J. Biomed. Mater. Res. 93B (2010) 520–530. [DOI] [PubMed] [Google Scholar]
- [683].Ma X, He Z, Han F, Zhong Z, Chen L, Li B, Colloids Surf. B: Biointerfaces 143 (2016) 81–87. [DOI] [PubMed] [Google Scholar]
- [684].Gurumurthy B, Tucci MA, Fan L, Benghuzzi HA, Pal P, Bidwell GL, Salazar Marocho SM, Cason Z, Gordy D, Janorkar AV, Adv. Healthcare Mater. 9 (2020) 1901385. [DOI] [PubMed] [Google Scholar]
- [685].Huang Z, Feng Q, Yu B, Li S, Mater. Sci. Eng. C 31 (2011) 683–687. [Google Scholar]
- [686].Zhou Y, Gu Z, Liu J, Huang K, Liu G, Wu J, Carbohydr. Polym. 230 (2020) 115640. [DOI] [PubMed] [Google Scholar]
- [687].Bhakta G, Lim ZXH, Rai B, Lin T, Hui JH, Prestwich GD, van Wijnen AJ, Nurcombe V, Cool SM, Acta Biomater. 9 (2013) 9098–9106. [DOI] [PubMed] [Google Scholar]
- [688].Holloway JL, Ma H, Rai R, Burdick JA, J. Control. Release 191 (2014) 63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [689].Kettenberger U, Luginbuehl V, Procter P, Pioletti Dominique P., J. Tissue Eng. Regen. Med. 11 (2017) 1974–1985. [DOI] [PubMed] [Google Scholar]
- [690].Liu LS, Thompson AY, Heidaran MA, Poser JW, Spiro RC, Biomaterials 20 (1999) 1097–1108. [DOI] [PubMed] [Google Scholar]
- [691].Yang X, Li Y, Liu X, Huang Q, He W, Zhang R, Feng Q, Benayahu D, Biomed. Mater. 12 (2016) 015001. [DOI] [PubMed] [Google Scholar]
- [692].Li H, Qi Z, Zheng S, Chang Y, Kong W, Fu C, Yu Z, Yang X, Pan S, Adv. Mater. Sci. Eng. 2019 (2019) e3027303. [Google Scholar]
- [693].Zhai P, Peng X, Li B, Liu Y, Sun H, Li X, Int. J. Biol. Macromol. 151 (2020) 1224–1239. [DOI] [PubMed] [Google Scholar]
- [694].Zhao W, Jin X, Cong Y, Liu Y, Fu J, J. Chem. Technol. Biotechnol. 88 (2013) 327–339. [Google Scholar]
- [695].Wakitani S, Goto T, Pineda SJ, Young RG, Mansour JM, Caplan AI, Goldberg VM, J. Bone Joint. Surg. Am. 76 (1994) 579–592. [DOI] [PubMed] [Google Scholar]
- [696].Wakitani S, Imoto K, Yamamoto T, Saito M, Murata N, Yoneda M, Osteoarthr. Cartil. 10 (2002) 199–206. [DOI] [PubMed] [Google Scholar]
- [697].Yamaoka H, Asato H, Ogasawara T, Nishizawa S, Takahashi T, Nakatsuka T, Koshima I, Nakamura K, Kawaguchi H, Chung U, Takato T, Hoshi K, J. Biomed. Mater. Res. 78A (2006) 1–11. [DOI] [PubMed] [Google Scholar]
- [698].Mimura T, Imai S, Okumura N, Li L, Nishizawa K, Araki S, Ueba H, Kubo M, Mori K, Matsusue Y, J. Biomed. Mater. Res. 98B (2011) 360–368. [DOI] [PubMed] [Google Scholar]
- [699].Ochi M, Uchio Y, Kawasaki K, Wakitani S, Iwasa J, J. Bone Jt. Surg. 84 (2002) 8. [DOI] [PubMed] [Google Scholar]
- [700].Dong M, Xu S, Bünger MH, Birkedal H, Besenbacher F, Adv. Eng. Mater. 9 (2007) 1129–1133. [Google Scholar]
- [701].Kilmer CE, Battistoni CM, Cox A, Breur GJ, Panitch A, Liu JC, ACS Biomater. Sci. Eng. 6 (2020) 3464–3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [702].Lu Z, Doulabi BZ, Huang C, Bank RA, Helder MN, Tissue Eng. Part A 16 (2010) 81–90. [DOI] [PubMed] [Google Scholar]
- [703].Ehlers E-M, Fuß M, Rohwedel J, Russlies M, Kühnel W, Behrens P, Ann. Anat. - Anat. Anz. 181 (1999) 513–518. [DOI] [PubMed] [Google Scholar]
- [704].Chung C, Beecham M, Mauck RL, Burdick JA, Biomaterials 30 (2009) 4287–4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [705].Kim IL, Khetan S, Baker BM, Chen CS, Burdick JA, Biomaterials 34 (2013) 5571–5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [706].Kwon MY, Wang C, Galarraga JH, Puré E, Han L, Burdick JA, Biomaterials 222 (2019) 119451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [707].Zhu D, Wang H, Trinh P, Heilshorn SC, Yang F, Biomaterials 127 (2017) 132–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [708].Chen F, Ni Y, Liu B, Zhou T, Yu C, Su Y, Zhu X, Yu X, Zhou Y, Carbohydr. Polym. 166 (2017) 31–44. [DOI] [PubMed] [Google Scholar]
- [709].Toh WS, Lim TC, Kurisawa M, Spector M, Biomaterials 33 (2012) 3835–3845. [DOI] [PubMed] [Google Scholar]
- [710].La Gatta A, Ricci G, Stellavato A, Cammarota M, Filosa R, Papa A, D’Agostino A, Portaccio M, Delfino I, De Rosa M, Schiraldi C, Int. J. Biol. Macromol. 103 (2017) 978–989. [DOI] [PubMed] [Google Scholar]
- [711].Yuan L, Li B, Yang J, Ni Y, Teng Y, Guo L, Fan H, Fan Y, Zhang X, Tissue Eng. Part A 22 (2016) 899–906. [DOI] [PubMed] [Google Scholar]
- [712].Liao E, Yaszemski M, Krebsbach P, Hollister S, Tissue Eng. 13 (2007) 537–550. [DOI] [PubMed] [Google Scholar]
- [713].Chen W-C, Wei Y-H, Chu I-M, Yao C-L, J. Tissue Eng. Regen. Med. 7 (2013) 665–672. [DOI] [PubMed] [Google Scholar]
- [714].Funayama A, Niki Y, Matsumoto H, Maeno S, Yatabe T, Morioka H, Yanagimoto S, Taguchi T, Tanaka J, Toyama Y, J. Orthop. Sci. 13 (2008) 225–232. [DOI] [PubMed] [Google Scholar]
- [715].Basalo IM, Chahine NO, Kaplun M, Chen FH, Hung CT, Ateshian GA, J. Biomech. 40 (2007) 1847–1854. [DOI] [PubMed] [Google Scholar]
- [716].Iovu M, Dumais G, du Souich P, Osteoarthr. Cartil. 16 (2008) S14–S18. [DOI] [PubMed] [Google Scholar]
- [717].Wang T, Lai JH, Yang F, Tissue Eng. Part A 22 (2016) 1348–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [718].Wang T, Lai JH, Han L-H, Tong X, Yang F, J. Mater. Chem. B 4 (2016) 7641–7650. [DOI] [PubMed] [Google Scholar]
- [719].Rogan H, Ilagan F, Yang F, Tissue Eng. Part A 25 (2019) 1404–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [720].Kim H, Lee Y, Kim Y, Hwang Y, Hwang N, Polymers 9 (2017) 655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [721].Liu Y, Zhou G, Cao Y, Engineering 3 (2017) 28–35. [Google Scholar]
- [722].Aisenbrey EA, Bryant SJ, Biomaterials 190–191 (2019) 51–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [723].Armiento AR, Stoddart MJ, Alini M, Eglin D, Acta Biomater. 65 (2018) 1–20. [DOI] [PubMed] [Google Scholar]
- [724].Tamaddon M, Wang L, Liu Z, Liu C, Bio-Des. Manuf. 1 (2018) 101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [725].Irawan V, Sung T-C, Higuchi A, Ikoma T, Tissue Eng. Regen. Med. 15 (2018) 673–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [726].Dinoro J, Maher M, Talebian S, Jafarkhani M, Mehrali M, Orive G, Foroughi J, Lord MS, Dolatshahi-Pirouz A, Biomaterials 214 (2019) 119214. [DOI] [PubMed] [Google Scholar]
- [727].Bowman S, Awad ME, Hamrick MW, Hunter M, Fulzele S, Clin. Transl. Med. 7 (2018) e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [728].Eming SA, Martin P, Tomic-Canic M, Sci. Transl. Med. 6 (2014) 265sr6–265sr6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [729].Ahn S, Yoon H, Kim G, Kim Y, Lee S, Chun W, Tissue Eng. Part C Methods 16 (2009) 813–820. [DOI] [PubMed] [Google Scholar]
- [730].Knapp DM, Barocas VH, Moon AG, Yoo K, Petzold LR, Tranquillo RT, J. Rheol. 41 (1998) 971. [Google Scholar]
- [731].Hartmann- Fritsch F, Biedermann T, Braziulis E, Luginbühl J, Pontiggia L, Böttcher-Haberzeth S, van Kuppevelt TH, Faraj KA, Schiestl C, Meuli M, Reichmann E, J. Tissue Eng. Regen. Med. 10 (2016) 81–91. [DOI] [PubMed] [Google Scholar]
- [732].Lotz C, Schmid FF, Oechsle E, Monaghan MG, Walles H, Groeber-Becker F, ACS Appl. Mater. Interfaces 9 (2017) 20417–20425. [DOI] [PubMed] [Google Scholar]
- [733].Ananta M, Brown RA, Mudera V, Tissue Eng. Part A 18 (2011) 353–361. [DOI] [PubMed] [Google Scholar]
- [734].Braziulis E, Diezi M, Biedermann T, Pontiggia L, Schmucki M, Hartmann-Fritsch F, Luginbühl J, Schiestl C, Meuli M, Reichmann E, Tissue Eng. Part C Methods 18 (2011) 464–474. [DOI] [PubMed] [Google Scholar]
- [735].Brown RA, Wiseman M, Chuo C-B, Cheema U, Nazhat SN, Adv. Funct. Mater. 15 (2005) 1762–1770. [Google Scholar]
- [736].Pensalfini M, Ehret AE, Stüdeli S, Marino D, Kaech A, Reichmann E, Mazza E, J. Mech. Behav. Biomed. Mater. 69 (2017) 85–97. [DOI] [PubMed] [Google Scholar]
- [737].Sohutskay DO, Buno KP, Tholpady SS, Nier SJ, Voytik-Harbin SL, Regen. Med. 15 (2020) 1295–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [738].Longaker MT, Chiu ES, Adzick NS, Stern M, Harrison MR, Stern R, Ann. Surg. 213 (1991) 292–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [739].Wu S, Deng L, Hsia H, Xu K, He Y, Huang Q, Peng Y, Zhou Z, Peng C, J. Biomater. Appl. 31 (2017) 1380–1390. [DOI] [PubMed] [Google Scholar]
- [740].Murphy SV, Skardal A, Song L, Sutton K, Haug R, Mack DL, Jackson J, Soker S, Atala A, Stem Cells Transl. Med. 6 (2017) 2020–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [741].Catanzano O, D’Esposito V, Acierno S, Ambrosio MR, De Caro C, Avagliano C, Russo P, Russo R, Miro A, Ungaro F, Calignano A, Formisano P, Quaglia F, Carbohydr. Polym. 131 (2015) 407–414. [DOI] [PubMed] [Google Scholar]
- [742].Qu J, Zhao X, Liang Y, Xu Y, Ma PX, Guo B, Chem. Eng. J. 362 (2019) 548–560. [Google Scholar]
- [743].Peattie RA, Nayate AP, Firpo MA, Shelby J, Fisher RJ, Prestwich GD, Biomaterials 25 (2004) 2789–2798. [DOI] [PubMed] [Google Scholar]
- [744].Peattie RA, Rieke ER, Hewett EM, Fisher RJ, Shu XZ, Prestwich GD, Biomaterials 27 (2006) 1868–1875. [DOI] [PubMed] [Google Scholar]
- [745].Xie Y, Upton Z, Richards S, Rizzi SC, Leavesley DI, J. Control. Release 153 (2011) 225–232. [DOI] [PubMed] [Google Scholar]
- [746].Huang J, Ren J, Chen G, Li Z, Liu Y, Wang G, Wu X, Mater. Sci. Eng. C 89 (2018) 213–222. [DOI] [PubMed] [Google Scholar]
- [747].Hui L, Chen Y, Cancer Lett. 368 (2015) 7–13. [DOI] [PubMed] [Google Scholar]
- [748].Albritton JL, Miller JS, Dis. Model. Mech. 10 (2017) 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [749].Polyak K, J. Clin. Invest. 121 (2011) 3786–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [750].Herrmann D, Conway JRW, Vennin C, Magenau A, Hughes WE, Morton JP, Timpson P, Carcinogenesis 35 (2014) 1671–1679. [DOI] [PubMed] [Google Scholar]
- [751].Misra S, Hascall VC, Markwald RR, Ghatak S, Front. Immunol. 6 (2015) 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [752].Bourguignon LYW, Int. J. Mol. Sci. 17 (2016) 517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [753].Xu S, Xu H, Wang W, Li S, Li H, Li T, Zhang W, Yu X, Liu L, J Transl. Med. 17 (2019) 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [754].Fang M, Yuan J, Peng C, Li Y, Tumour Biol. 35 (2014) 2871–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [755].Schwertfeger KL, Cowman MK, Telmer PG, Turley EA, McCarthy JB, Front. Immunol. 6 (2015) 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [756].Sato N, Kohi S, Hirata K, Goggins M, Cancer Sci. 107 (2016) 569–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [757].Seufferlein T, Ducreux M, Hidalgo M, Prager G, Cutsem EV, Eur. Oncol. Haematol. 14 (2018) 40. [Google Scholar]
- [758].Siegel RL, Miller KD, Jemal A, CA CANCER J. CLIN. 68 (2018) 7–30. [DOI] [PubMed] [Google Scholar]
- [759].North CM, Christiani DC, Semin. Thorac. Cardiovasc. Surg. 25 (2013) 87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [760].Jin X, Mu P, Breast Cancer (Auckl) 9 (2015) 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [761].Wu W, Chen L, Wang Y, Jin J, Xie X, Zhang J, Medicine 99 (2020) e20438. [DOI] [PubMed] [Google Scholar]
- [762].Bertrand P, Girard N, Delpech B, Duval C, D’Anjou J, Dauce JP, Int. J. Cancer 52 (1992) 1–6. [DOI] [PubMed] [Google Scholar]
- [763].Bonnesœur S, Morin- Grognet S, Thoumire O, Cerf DL, Boyer O, Vannier J-P, Labat B, J. Biomed. Mater. Res. Part A 108 (2020) 1256–1268. [DOI] [PubMed] [Google Scholar]
- [764].Ondeck MG, Kumar A, Placone JK, Plunkett CM, Matte BF, Wong KC, Fattet L, Yang J, Engler AJ, Proc. Natl. Acad. Sci. U. S. A. 116 (2019) 3502–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [765].Wang Y, Mirza S, Wu S, Zeng J, Shi W, Band H, Band V, Duan B, Oncotarget 9 (2018) 32191–32203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [766].Campbell JJ, Davidenko N, Caffarel MM, Cameron RE, Watson CJ, PLoS ONE 6 (2011) e25661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [767].Mazio C, Casale C, Imparato G, Urciuolo F, Netti PA, Acta Biomater. 73 (2018) 236–249. [DOI] [PubMed] [Google Scholar]
- [768].Libring S, Shinde A, Chanda MK, Nuru M, George H, Saleh AM, Abdullah A, Kinzer-Ursem TL, Calve S, Wendt MK, Solorio L, Cancers 12 (2020) 1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [769].Shinde A, Libring S, Alpsoy A, Abdullah A, Schaber JA, Solorio L, Wendt MK, Mol. Cancer Res. 16 (2018) 1579–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [770].Evanko SP, Potter-Perigo S, Petty LJ, Workman GA, Wight TN, Matrix Biol. 42 (2015) 74–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [771].Otranto M, Sarrazy V, Bonté F, Hinz B, Gabbiani G, Desmouliere A, Cell Adhes. Migr. 6 (2012) 203–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [772].Gioiella F, Urciuolo F, Imparato G, Brancato V, Netti PA, Adv. Healthc. Mater. 5 (2016) 3074–3084. [DOI] [PubMed] [Google Scholar]
- [773].Brännvall K, Bergman K, Wallenquist U, Svahn S, Bowden T, Hilborn J, Forsberg-Nilsson K, J. Neurosci. Res. 85 (2007) 2138–2146. [DOI] [PubMed] [Google Scholar]
- [774].Zhang H, Wei Y, Tsang K, Sun C, Li J, Huang H, Cui F, An Y, J. Transl. Med. 6 (2008) 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [775].Rao SS, DeJesus J, Short AR, Otero JJ, Sarkar A, Winter JO, ACS Appl. Mater. Interfaces 5 (2013) 9276–9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [776].Kuo Y-C, Hsueh C-H, Mater. Sci. Eng. C 76 (2017) 760–774. [DOI] [PubMed] [Google Scholar]
- [777].Xu W, Hu R, Fan E, Han D, Ann. Otol. Rhinol. Laryngol. 120 (2011) 123–130. [DOI] [PubMed] [Google Scholar]
- [778].Chen F, Le P, Lai K, Fernandes-Cunha GM, Myung D, Chem. Mater. 32 (2020) 5208–5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [779].Gansau J, Buckley CT, J. Funct. Biomater. 9 (2018) 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [780].Segura T, Chung PH, Shea LD, Biomaterials 26 (2005) 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [781].Harrington S, Williams J, Rawal S, Ramachandran K, Stehno-Bittel L, Tissue Eng. Part A 23 (2017) 1088–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]