Figure 2.
Sequencing data properties
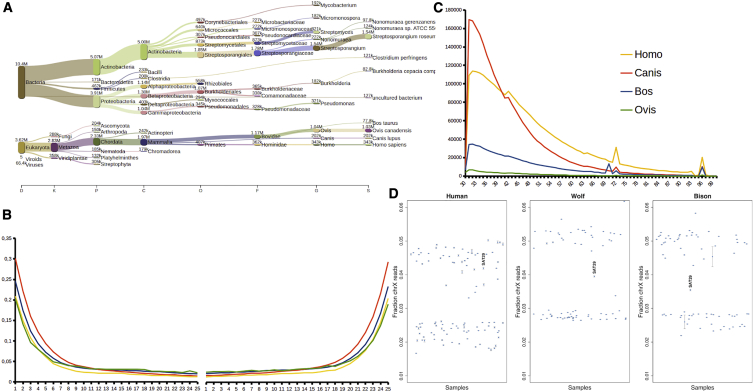
(A) Metagenomic prediction with centrifuge.
(B) Deamination patterns for the filtered reads mapped against the four reference genomes: Ovis aries, Homo sapiens, Bos taurus, and Canis lupus.
(C) Fragment length distributions for the four aligned mammalian species.
(D) X chromosome read proportions. For each of the three species, the fraction of nuclear reads mapping to the X chromosome is displayed for the SAT29 sample as well as a number of previously published genomes for comparison. For wolf and bison, the comparative genomes are the modern and ancient genomes used for the ancestry analyses, while for human they are 101 ancient genomes from a study of the Eurasian steppe.18 Bars denote 95% binomial confidence intervals.19
See also Figures S1 and S6 and Data S1.