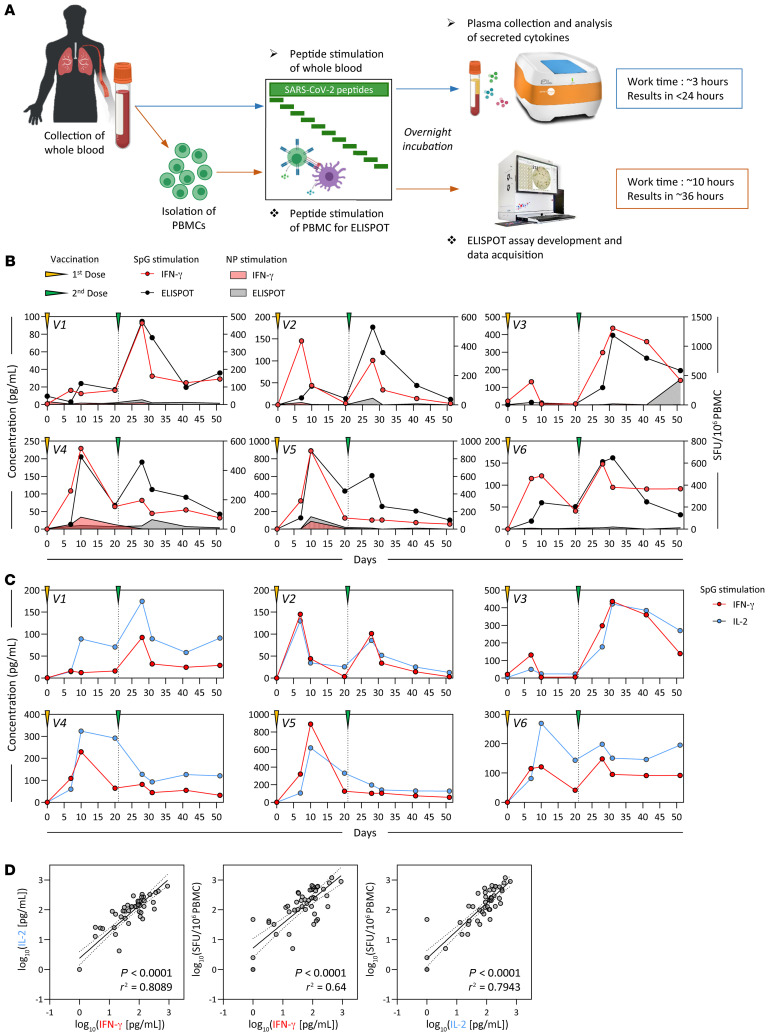
Figure 1. Detection of SARS-CoV-2 spike–specific T cells by peptide stimulation of whole peripheral blood from vaccinated individuals.
(A) Schematic representation of the workflow for the direct peptide stimulation of whole peripheral blood and the subsequent detection of cytokine secretion compared with a standard IFN-γ ELISPOT assay. (B) Six healthy individuals were vaccinated with 2 doses of BNT162b2 according to the recommended schedule (21 days apart), and whole-blood samples were longitudinally analyzed 7, 10, 20, and 30 days after each dose. The collected whole blood was either directly stimulated for 16 hours with peptide pools specific for the spike protein (red or black line) or NP (red or black shaded area), or immediately processed with Ficoll density gradient centrifugation to isolate PBMCs. A standard IFN-γ ELISPOT assay using the SpG- or NP-specific peptide pools was then set up using the freshly isolated PBMCs. The quantity of secreted IFN-γ in stimulated whole blood (red line) was compared with the frequency of peptide-reactive PBMCs quantified by IFN-γ ELISPOT (black line). (C) The levels of secreted IL-2 (blue line) in whole blood stimulated with the SpG peptide pool were compared with the amount of IFN-γ detected. (D) Linear regression analysis of the concentrations of IFN-γ and IL-2 in SpG-specific peptide pool–stimulated whole blood and the corresponding frequency of spike-specific PBMCs (n = 6; 48 samples). Dotted lines denote the 95% CI.