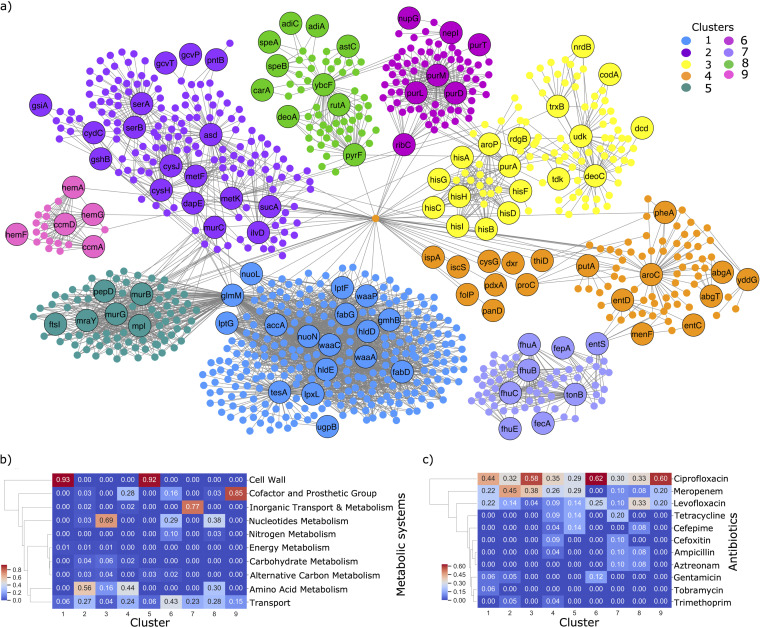
FIG 6.
Effect of genetic determinants on reaction fluxes. (a) Bipartite network with genes and reactions as nodes. Labeled nodes represent the genes, whereas unlabeled nodes represent reactions. A gene and reaction are connected by an edge if the deletion of the gene reduces the reaction flux by at least 10%. Genes and reactions are highlighted according to the cluster they were assigned to via the Networkx modularity algorithm. Note that to reduce the initial size of the network, we only included clusters of size greater than 10. (b) Heatmap showing the metabolic systems associated with each of the nine clusters. A gene was associated with a metabolic system, if the flux span of at least one reaction correlated with the system was reduced after the gene was deleted. (c) Heatmap showing the antibiotics associated with each cluster. Genes occurring in multiple antibiotics were accounted for twice. Hierarchical clustering was applied to the rows of each heatmap (metabolic systems or antibiotic class) using the single linkage method and Euclidean distance as the metric. The gene counts have also been normalized by the total number of genes in each cluster in each heatmap.