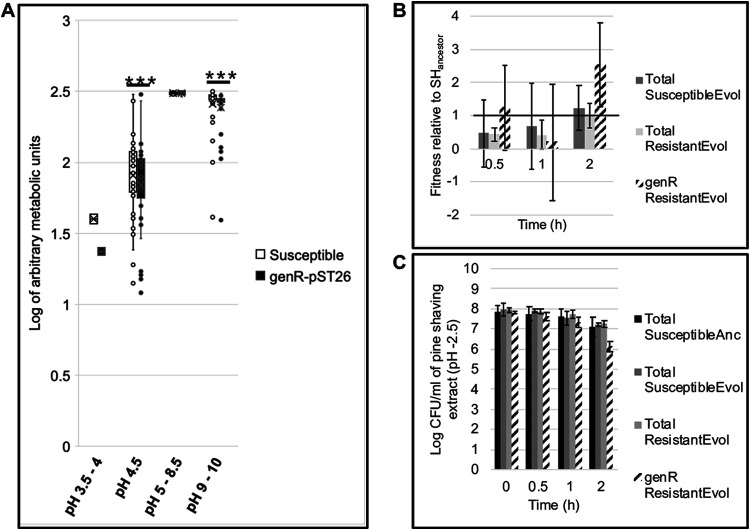
FIG 6.
S. Heidelberg exposure to acidic pH imposes negative frequency-dependent selection for IncI1. (a) Box plot comparing the metabolic activity of one evolved susceptible (no IncI1) and one genR (carries p2ST26) S. Heidelberg strain using phenotype microarray (PM) plates (Biolog Inc.). Preconfigured PM plates are composed of microtiter plates with one negative-control well and 95 wells prefilled with or without nitrogen and at pH of 3.5 to 10 (n = 1 plate per strain; the number of wells per plate for pH 3.5 to 4, 4.5, 5 to 8.5, and 9 to 10 was 2, 6, 37, and 38, respectively; error bars, standard deviation; ***, P < 0.001 (Wilcoxon signed-rank test). (b and c) Fitness and abundance of evolved susceptible and genR S. Heidelberg strains when exposed to pine shaving extract of pH 2.5. Each bar represents the average fitness or abundance (CFU per ml of pine shaving extract) of three individual populations that were established from three single bacterial colonies from three different strains (error bars, standard deviation; P > 0.05; Kruskal-Wallis rank-sum test). The horizontal line in b represents the fitness of three S. Heidelberg ancestor populations established from three single bacterial colonies. (Anc, ancestor; Evol, evolved, i.e., S. Heidelberg isolate recovered during in vivo experiment; genR, population resistant to gentamicin). Values of <1 show a reduction in fitness, while >1 indicates an increase in fitness.