Figure 2.
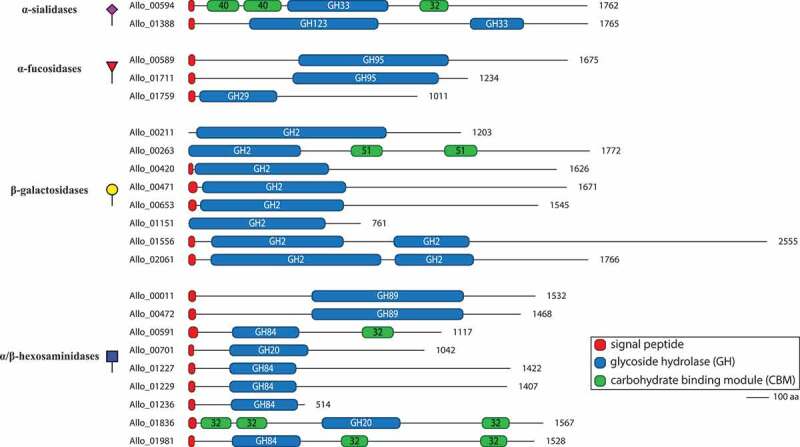
Domain architecture of putative mucin O-glycan targeting CAZymes in genome of A. mucolyticum. CAZymes are divided in groups based upon their predicted enzyme activity: from top to bottom, α-sialidases, α-fucosidases, β-galactosidases and α/β-hexosaminidases. The displayed domains are those identified by HMMER and SignalP 4.0 and are drawn to scale with the size of the polypeptide backbone, in number of amino acids, indicated at the right side of each protein
