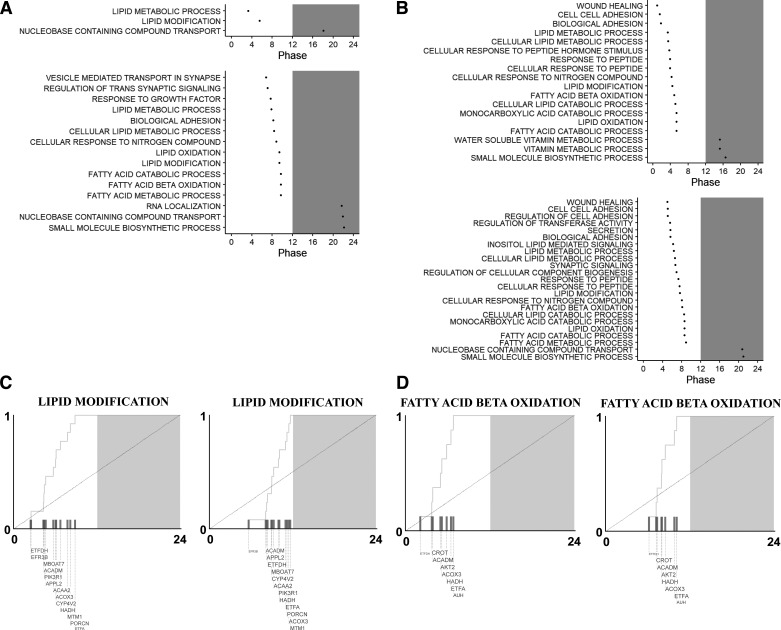
Figure 6.
Phase set enrichment analysis demonstrates preferential shifts in the expression of metabolic pathways. Phase set enrichment analysis (PSEA) for male (A) and female (B) normal circadian (NC; A and B, top) and chronic jetlag (CJ; A and B, bottom). All stably rhythmic genes in the transcriptome were entered into the PSEA JAVA script tool along with their respective phase (time of peak expression). Pathways were examined for differences from a uniform background based on the Kuiper test. Statistically significant (q < 0.05) gene ontology (GO) biologic process (BP) pathways are shown in A and B along with their respective phase. Shading represents the period of darkness [ZT12-ZT24 (zeitgeber time)]. Pathways were similar between NC and CJ conditions. Many metabolism-related pathways, especially lipid metabolism pathways, were present in both NC and CJ conditions and underwent a phase shift. Selected pathways are shown for both males (C) and females (D). Contributing genes for each pathway are shown as space allows. Gene font size is proportional to the magnitude of the contribution of a gene to each pathway.