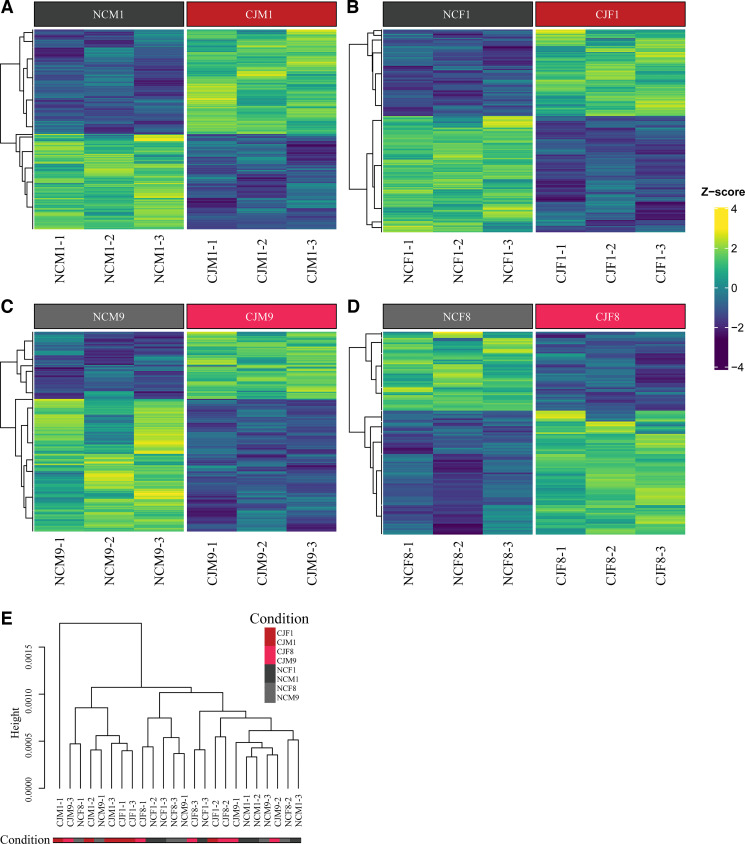
Figure 11.
Bulk RNA-sequencing following chronic jetlag reveals persistent pancreatic differential gene expression despite normalization. Heatmaps were created following differential gene expression testing with edgeR. All significantly different genes (q < 0.05) were included. Results are shown for males (A and C) and females (B and D) during early {day 1 [zeitgeber time (ZT) 8]; Fig. 11, A and B} and late [day 8/9 (ZT8); C and D] normalization. Three mice were assessed for each time point and condition. The data demonstrate that significant differences between normal circadian and chronic jetlag mice exist even after 8/9 days of normalization. Hierarchical clustering (E) was performed on the Euclidean distances computed between samples. With few exceptions, chronic jetlag mice on day 1 exhibited transcriptomic signatures that clustered separately from normal circadian mice. [NCM1 = normal circadian male day 1 of normalization (zeitgeber time 8), NCM9 = normal circadian male day 9 of normalization (ZT8), NCF1 = normal circadian female day 1 of normalization (ZT8), NCF8 = normal circadian female day 8 of normalization (ZT8), CJM1 = chronic jetlag male day 1 of normalization (ZT8), CJM9 = chronic jetlag male day 9 of normalization (ZT8), CJF1 = chronic jetlag female day 1 of normalization (ZT8), CJF8 = chronic jetlag female day 8 of normalization (ZT8)].