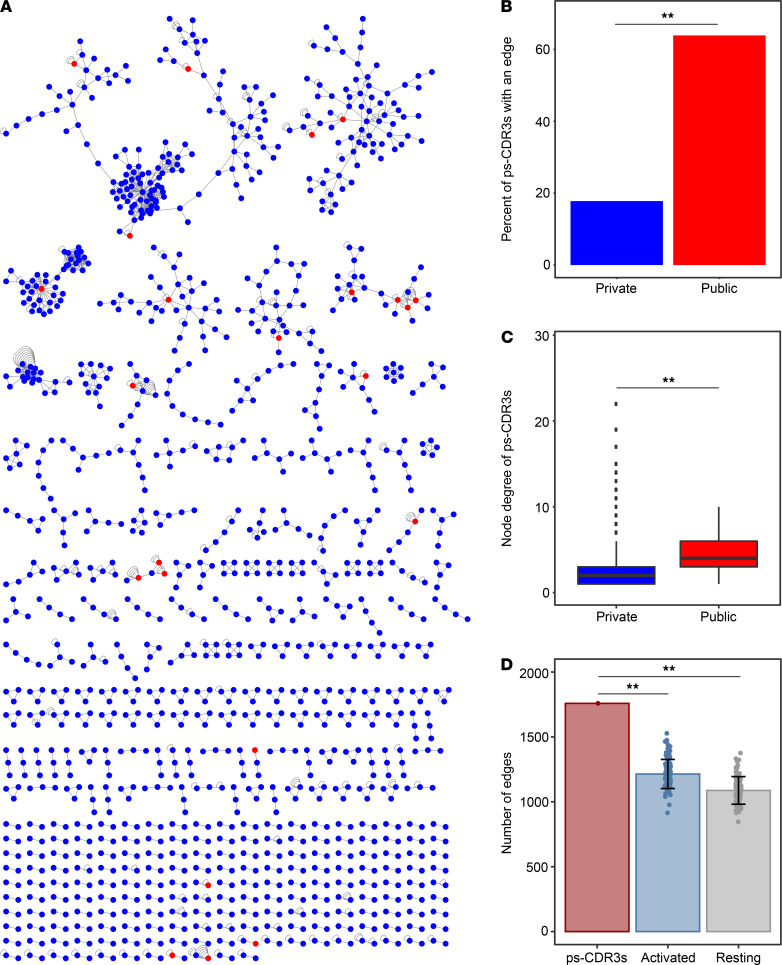
Figure 5. Network analysis reveals that public putatively peanut-specific CDR3β sequences are core sequences with more structure than activated and resting CDR3βs.
(A) Network analysis of putatively peanut-specific CDR3β sequences (ps-CDR3s), where each node represents a unique ps-CDR3 amino acid sequence. Edges were created between the nodes if these were within a Levenshtein distance of 1 or if the nodes shared an enriched motif. Self-edges were created on nodes to represent additional unique nucleotide sequences encoding the same amino acid sequence (i.e., convergence). Blue nodes represent private ps-CDR3s, and red nodes represent public ps-CDR3s. (B) The percentage of public ps-CDR3s with an edge to another ps-CDR3 (as shown in A) was higher than that of private ps-CDR3s (**P < 0.01, Fisher’s exact test). (C) The overall node degree (number of edges per node) of public ps-CDR3s was higher than that of private ps-CDR3s. Box plots represent distribution of the node degree of all private (blue) and public (red) ps-CDR3s in the graph (**P < 0.01, Mann-Whitney U test). (D) The number of total edges created among the ps-CDR3s compared with the median number of edges created among 100 equal-sized random resamplings of total activated and resting CDR3βs. Error bars represent standard deviation (**P < 0.01, Z test).