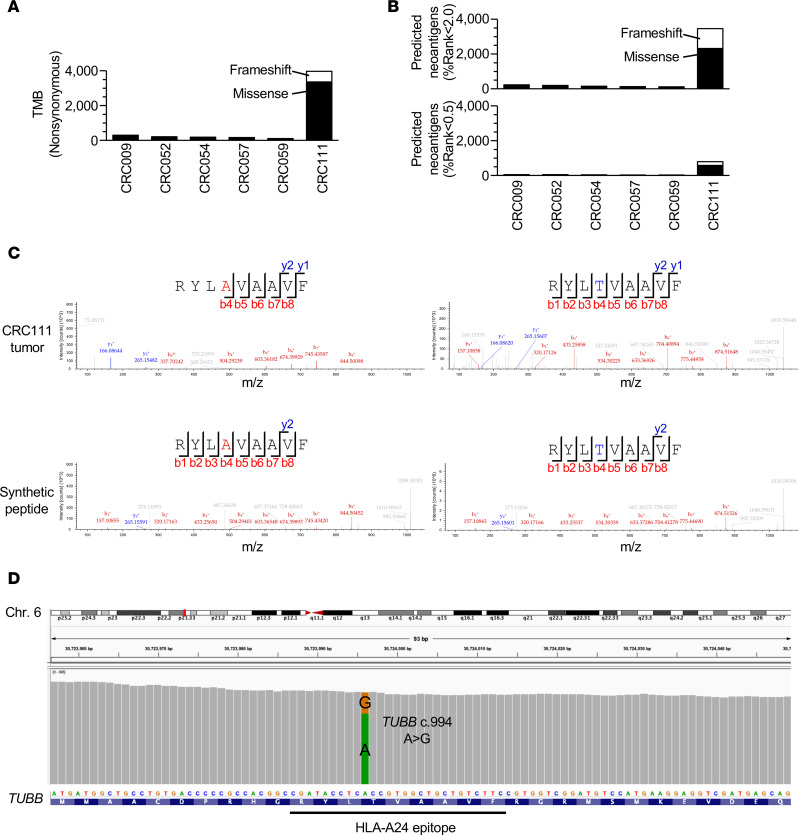
Figure 3. Proteogenomic identification of a neoantigen directly from dMMR-CRC tissue.
(A) Numbers of nonsynonymous (missense and frameshift) somatic mutations in CRC tumor tissues with pMMR (CRC009, -052, -054, -057, and -059) and dMMR (CRC111). (B) Numbers of neoantigens predicted in silico using WES and RNA-seq data under the condition of NetMHCpan-4.1 %rank < 2.0 or < 0.5, the TPM of the source genes > 0 and the peptide length of 8–11 amino acids. (C) Neoantigen and its WT counterpart sequences identified in CRC111 tumor tissue using a proteogenomic approach at an FDR of 0.01. MS/MS spectra and the corresponding b- and y-fragment ions of both endogenous and synthetic peptides are shown. (D) Coverage track of the TUBB gene mutation in CRC111 tumor tissue visualized using the Integrative Genomics Viewer (IGV) (http://software.broadinstitute.org/software/igv/).