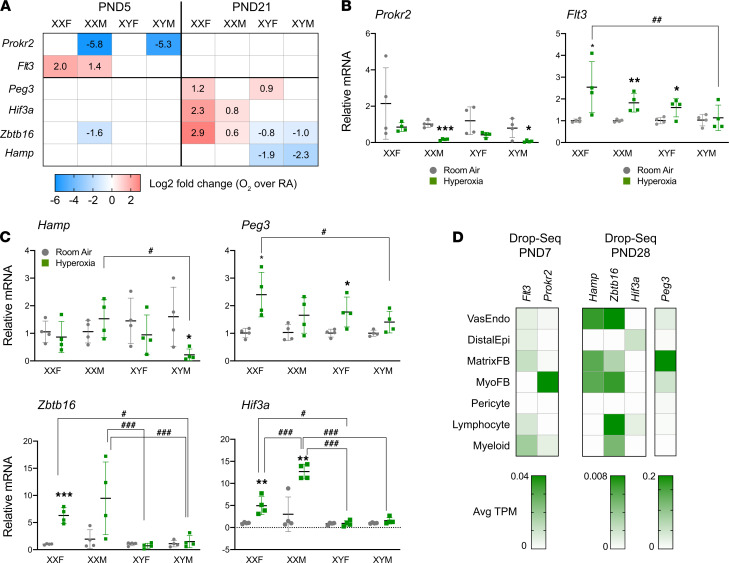
Figure 10. Independent validation of RNA-Seq results for selected genes with qPCR and cell-specific expression in neonatal mice.
A subset of highly expressed differentially expressed genes were validated in an independent cohort of FCG neonatal mice. (A–C) The fold change of the selected genes in the RNA-Seq experiment are shown in A, and the changes in the qPCR experiments are shown for DEGs at P5 (Flt3 and Prokr2) (B) and for DEGs at P21 (Hamp, Peg3, Zbtb16 and Hif-3α) (C). Data are shown as mean ± SD from 4 individual animals. Analysis done by 3-way ANOVA to assess the effect of treatment, chromosomal sex, and gonadal sex, as well as the interactions between the independent variables. Significant differences between room air and hyperoxia within genotype are indicated by *P < 0.05, **P < 0.01, and ***P <0.001. Significant differences between hyperoxia-exposed mice between different genotypes are indicated by #P < 0.05, ##P < 0.01, and ###P < 0.001. (D) For these selected genes, we also evaluated the expression in publicly available neonatal lung single-cell expression database (LungMAP).