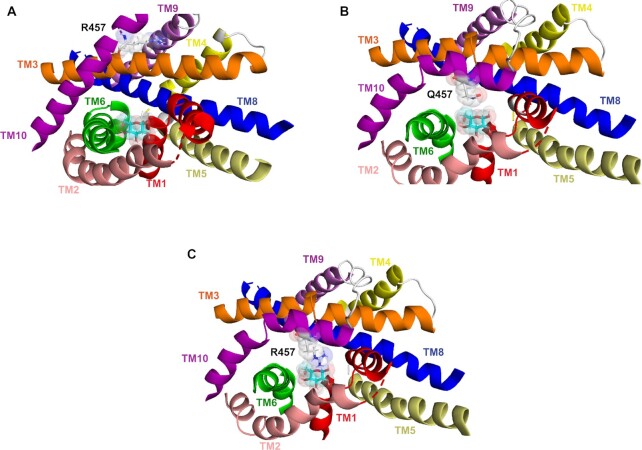
Figure 10.
Molecular model showing sugar-binding site of WT hSGLT1 and mutant Q457R. The molecular models of the outward and inward-facing conformations of hSGLT1 were based on the crystal structures of the sodium sialic acid symporter SiaT from Proteus mirabilis (PDB ID 5NV9) and the sodium glucose cotransporter vSGLT from Vibrio parahaemolyticus (PDB ID 3DH4).34 Computational and experimental approaches were used to validate the homology models. A sugar-binding site in outward open conformation. The sugar coordinating residues Q457, H83, Y290, N78, and E102 are not shown. Note that Q457R does not interact with sugar in this conformation. B. The sugar-binding site in the inward conformation. Note that the inward tilt of TM10 positions Q457 to interact with the pyranose oxygen and the C#6 –OH of glucose. C. Sugar binding site for mutant Q457R in the inward conformation. Note that in this conformation the side-chain of Arg457 encounters steric hindrance to binding with glucose.