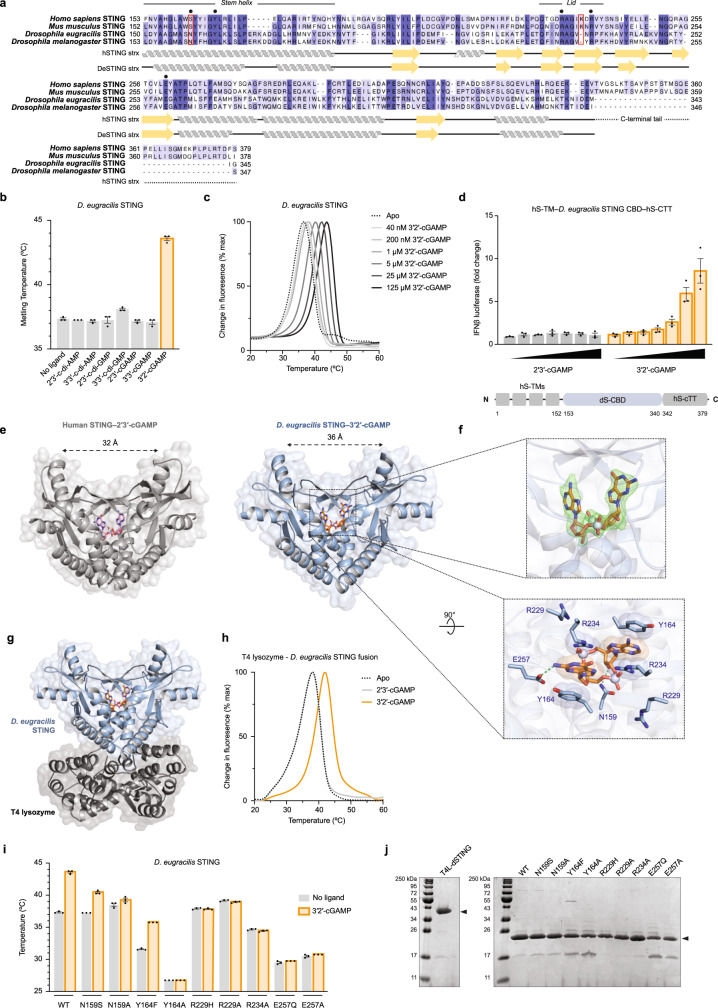
Extended Data Fig. 8. Structural and biochemical analysis of dSTING.
a, Alignment of the C-terminal CDN-binding domains of human STING, mouse STING, D. eugracilis STING and D. melanogaster STING. Architecture of the core CDN-binding domain is conserved across metazoans; the disordered C-terminal tail, which controls IRF3–IFNβ signalling, is specific to vertebrates8,21. Ligand-interacting residues selected for mutational analysis are denoted with a black circle; Diptera-specific adaptations are highlighted with a red outline. All structural and biochemical experiments were performed with a D. eugracilis STING construct terminating at I340. b, In vitro thermal denaturation assay analysing dSTING interactions with a panel of CDNs. Only 3′2′-cGAMP forms a thermostable complex with dSTING in vitro (see also Fig. 3d). 2′3′-cGAMP is known to be capable of stimulating dSTING-dependent signalling in vivo26, supporting that dSTING can engage with 2′3′-cGAMP with lower affinity. This observation is consistent with the weaker recognition of bacteria-derived 3′3′-cGAMP and 3′3′-c-di-GMP by human STING2,4. c, In vitro thermal denaturation assay demonstrating concentration-dependent thermal shift induced by 3′2′-cGAMP. d, Dose titration of 2′3′-cGAMP and 3′2′-cGAMP in human cells demonstrating selective response by dSTING to 3′2′-cGAMP. The D. eugracilis CDN-binding domain (CBD) was adapted for downstream signalling in human cells by addition of N-terminal human transmembrane (hTM) domains and the human C-terminal tail (hCTT). e, Comparison of the human STING–2′3′-cGAMP and dSTING–3′2′-cGAMP crystal structures reveals a conserved closed homodimer architecture in which apical ‘wings’ are spread 32–36 Å, demonstrating high-affinity engagement with an endogenous ligand. f, Enlarged cutaways of 3′2′-cGAMP in the dSTING crystal structure. Above: the simulated annealing FO−FC omit map (contoured at 3 σ). Below: a top-down view highlighting key dSTING–3′2′-cGAMP contacts. g, Full crystal structure used to determine the structure of D. eugracilis STING in complex with 3′2′-cGAMP. T4 lysozyme is fused to the N terminus of the D. eugracilis STING CBD. h, Thermal denaturation assay as in Fig. 3d demonstrating that N-terminal fusion of T4 lysozyme does not impair dSTING recognition of 3′2′-cGAMP. i, Mutational analysis of key ligand-interacting residues in dSTING; the thermal denaturation assay was used to analyse 3′2′-cGAMP recognition. Mutations that conserve functional contacts with 3′2′-cGAMP (Y164F) maintain ligand recognition; mutations that ablate contacts abrogate ligand binding. N159S exhibits diminished ability to recognize 3′2′-cGAMP. Data in b and i are mean ± s.e.m. of the average Tm calculated from n = 2 technical replicates in n = 3 independent experiments. Data in c are representative of n = 3 independent experiments. Data in d are mean ± s.e.m. of n = 3 technical replicates and representative of n = 3 independent experiments. j, SDS–PAGE and Coomassie stain analysis of purified WT and mutant proteins.