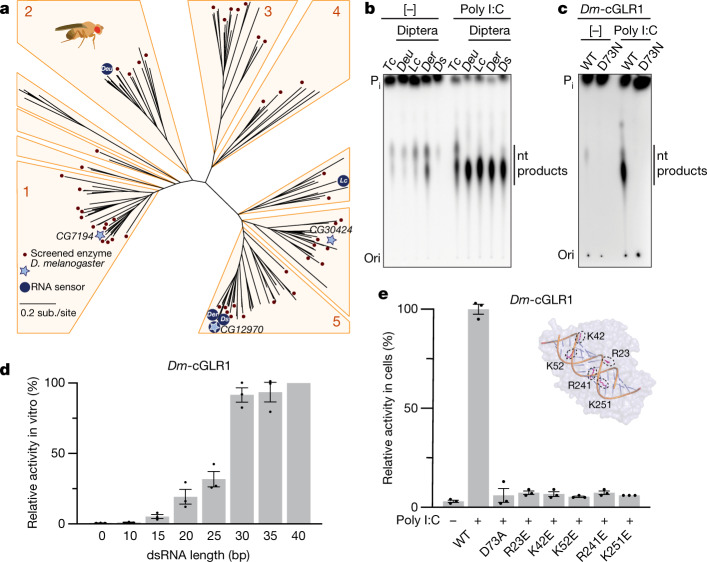
Fig. 2. Drosophila cGLR1 senses long dsRNA.
a, Phylogeny representing 153 Diptera cGLR genes clustered into clades 1–5 (less than 30% sequence identity between clades). Forty-one of forty-two analysed Diptera species encode enzymes in clade 5, including D. melanogaster CG12970 (cGLR1) and CG30424. Enzymes analysed by forward biochemical screen (red dot) and identified as dsRNA-sensing cGLRs (blue circle) are denoted. b, Diptera cGLRs identified in the screen are activated to form a nucleotide product by the dsRNA mimic poly I:C. c, Mutation to the Dm-cGLR1 active site ablates enzymatic activity. Data in b and c are representative of n = 3 independent experiments. WT, wild type. d, Dm-cGLR1 in vitro activity was monitored in the presence of a panel of dsRNAs and quantified relative to 40 bp dsRNA. Data are the mean ± s.e.m. of n = 3 independent experiments. e, Analysis of Dm-cGLR1 activity in human cells using mammalian STING and IFNβ reporter induction, quantified relative to WT activity. Dm-cGLR1 signalling in cells is dependent on stimulation of dsRNA and mutation of the catalytic site, or the predicted ligand-binding residues ablates activity. Data are mean ± s.e.m. of n = 3 technical replicates and representative of n = 3 independent experiments. The inset shows a model of the Tc-cGLR–dsRNA complex based on the hcGAS–dsDNA structure (PDB: 6CTA)14 used to predict the Dm-cGLR1 ligand-binding residues R23, K42, K52, R241 and K251.