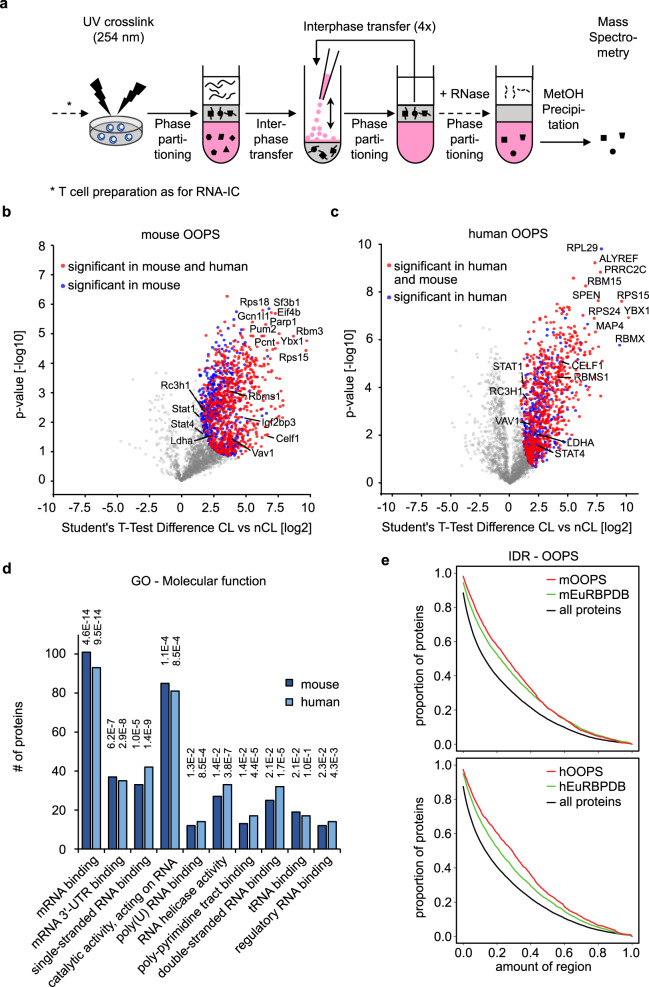
Fig. 3. The global RNA-bound proteome of T helper cells.
TH0 cultures from three mice or four human donors were used as biologic replicates (n = 3 or n = 4) to characterize proteins interacting with RNA. a Schematic overview of the OOPS method29 with an increased number (5) of phase partitioning cycles. b and c Volcano plots from two-sided Student’s T-test analysis using a permutation-based FDR method for multiple hypothesis corrections showing the −log10 p-value plotted against the log2 fold-change comparing the organic phase after RNase treatment of the interphase of OOPS experiments of the crosslinked mouse (b) or human CD4+ T cells (c) versus the non-crosslinked sample. Red dots represent proteins significant at a 5% FDR cutoff level in both mouse and human OOPS experiments and blue dots represent proteins significant only in mice or humans, respectively. d Enrichment analysis of GO molecular function terms of significant proteins in mouse or human OOPS data. Enriched terms are depicted and were calculated as described for RNA-capture data in Fig. 2d. e Distribution of intrinsically disordered regions in all Uniprot reviewed protein sequences (black line), in proteins in the mouse EuRBPDB database (green), and in proteins significant in the mouse OOPS data (red line). The same plot is shown for human data at the bottom. According to two-sided Kolmogorov–Smirnov testing, the IDR distribution differences between RNA-IC (red lines) and all proteins (black lines) are highly significant in mice and man and reach the smallest possible p-value (p < 2.2 × 10−16).