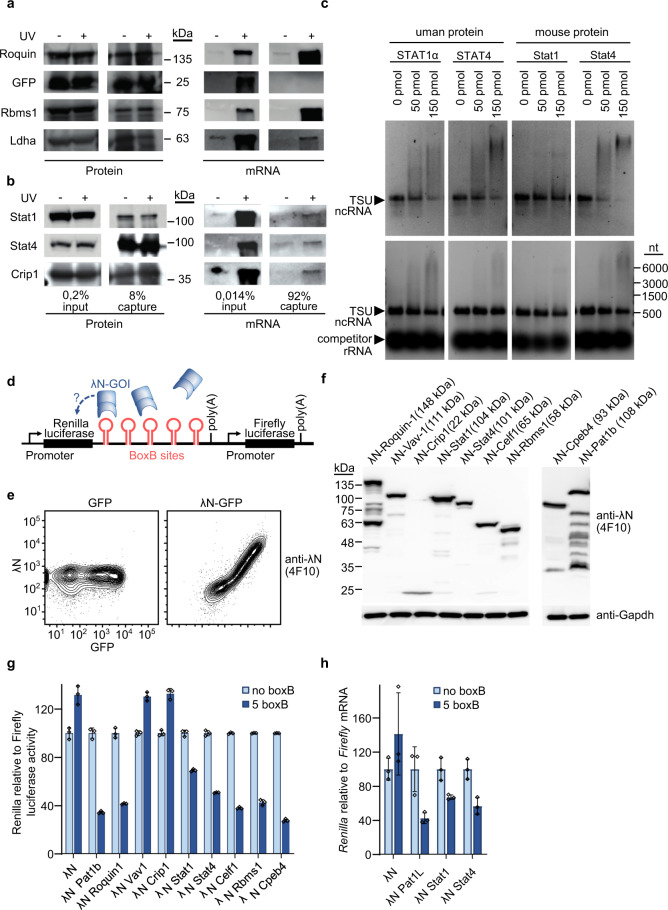
Fig. 5. Stat1 and Stat4 bind to RNA and have regulatory potential.
Transfected or recombinant Stat1 and Stat4 were analyzed for interaction with RNA and regulatory potential. a, b Semi-quantitative identification method for RBPs as described in75. In short, GFP-fused proteins were transfected into 293T cells, crosslinked or left untreated, and subsequently immunoprecipitated using an anti-GFP antibody. The obtained samples were divided for protein and RNA detection after transfer to membranes (n = 2). c Electrophoretic mobility shift assays using purified human STAT1α and STAT4 as well as mouse Stat1 and Stat4 proteins to bind in vitro-transcribed non-coding RNA ‘TSU' (481 nt) in the absence (upper panels) and presence (lower panels) of competitor RNA (baker’s yeast rRNA) (n = 2). d Schematic representation of the tethering assay that was used to investigate a possible influence of the genes of interest (GOI) on renilla luciferase expression. The affinity of the λN peptide targets the respective fusion protein to boxB stem-loop structures (5x) in the 3′-UTR of a Renilla luciferase gene where it can exert its function, if exciting. e FACS plots using the new monoclonal antibody 4F10 to demonstrate specific λN detection of a λN-GFP fusion protein expressed in 293T cells. f Western blot showing the expression of the indicated λN-GOI proteins in 293T cells after transfection (n = 2). g Tethering assay results performed in HeLa cells as described in (d). Two negative controls were implemented, using constructs without boxB sites or λN expression without fusion to a GOI. Each measurement was performed in triplicate and was independently repeated at least twice (n = 2). Error bars of three technical replicates, mean ± s.d. h Experiment performed as in (g), however, Renilla and Firefly RNA levels were assessed by qPCR (n = 3). Error bars of biological replicates, mean ± s.d. Source data are provided as a Source Data file.