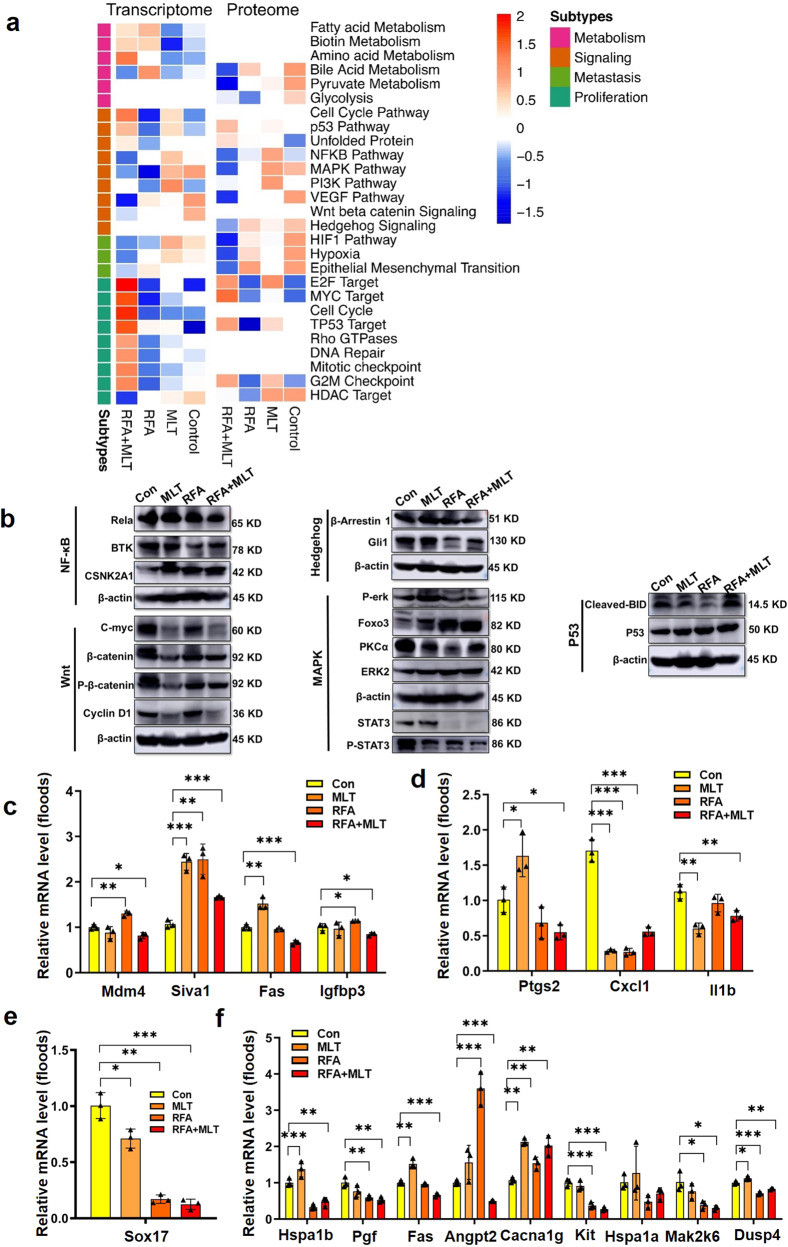
Fig. 5.
Combined treatment with RFA and MLT reduced tumor malignancy in non-ablated areas, altered tumor metabolism, and the tumor microenvironment. a The integrative multi-omics analysis of cancer proliferation and malignancy-related pathways. Heatmap of alteration pathways in the transcriptomic and proteomic subtypes. Left, alteration pathways identified from the transcriptome. Right, alteration pathways are identified from the proteome. Color of each cell represents the average ssGSEA enrichment scores of that subtype; red denotes activation and blue denotes inhibition. Blank cells represent non-enrichment. b The validation of cancer proliferation and malignancy-related pathways. Western blot analysis of P53 (Cleaved-BID and P53), Wnt (C-myc, β-catenin, P-β-catenin, and Cyclin D1), Hedgehog (β-Arrestin 1 and Gli1), MAPK (P-erk, Foxo3, PKC, ERK2, p-STAT3 and STAT3), and NFкB (Rela, BTK, CSNK2A1) pathway-altered proteins in the RFA + MLT, RFA, MLT, and Control groups; The normalized altered gene expression of c P53 (Mdm4, Siva1, Fas, and Igfbp3), d NFкB (Ptgs2, Cxcl1, and Il1b), e Wnt (Sox17), and f MAPK (Hspa1b, Pgf, Fas, Angpt2, Cacna1g, Kit, Hspa1a, Map2k6, Dusp4, and Il1b) signal pathways in the RFA + MLT, RFA, MLT, and Control groups (*P < 0.05; **P < 0 .01; ***P < 0. 001)