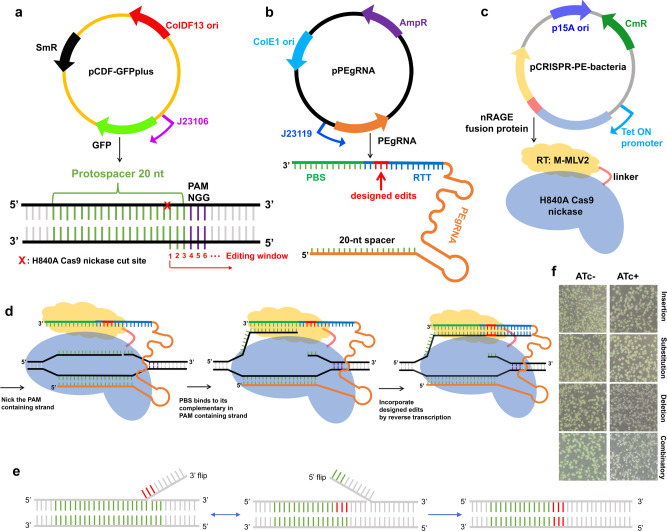
Fig. 1. A three-plasmid system for evaluation of CRISPR-Prime Editing system in E. coli.
a The plasmid map of the reporter vector pCDF-GFPplus, which carries a constitutive promoter J23106 driving expression of fast folding GFP. The plasmid contains a spectinomycin-resistance (SmR) gene, and the ColDF13 origin (ori). An illustration of the target DNA composition is shown below the plasmid map. b The plasmid map of the PEgRNA transcript bearing vector, which contains the ColE1 ori and an ampicillin-resistance (AmpR) gene for selection. The PEgRNA transcript is under control of the constitutive promoter J23119. A detailed structure is shown beneath, the 3′ extension sequence is composed of a PBS (in green) and a RTT (in blue), which carries the intended edits (in red). c Plasmid map of the CRISPR-PE-bacteria vector, which carries a p15A ori and a chloramphenicol-resistance gene (CmR) for selection. The E. coli codon optimized, tetracycline inducible promoter driven Cas9n-M-MLV2 fusion protein consists of a H840A Cas9 nickase (Cas9n), a 33-aa flexible linker, and a moloney murine leukemia virus (M-MLV) variant M-MLV2, described previously19 with the following mutations: D200N, L603W, T306K, W313F, and T330P compared to the WT M-MLV (GenBank: AAC82568.2). d A schematic model for DNA engineering with CRISPR-Prime Editing system for E. coli. After being expressed, the Cas9n-M-MLV2:PEgRNA complex binds to the targeted DNA sequence in a sgRNA-dependent and PAM-dependent manner. The Cas9n domain within the fusion protein nicks the PAM-containing strand, freeing the adjacent DNA sequence. Subsequently, this piece of single stranded DNA hybridizes to the PBS, then primes reverse transcription of new DNA containing the designed edits based on the RTT within the 3′-extension of the PEgRNA transcript. e Two possible consequences of CRISPR-Prime Editing. It normally has an equilibration between the edited 3′ flap and the unedited 5′ flap, only the cleavage of the 5′ flap leads to the desired editing. f Colony views of E. coli strains transformed with CRISPR-Prime Editing systems carrying designed edits of TAA insertion, T to A substitution, T deletion and the combinatorial edits with and without 200 ng/mL ATc induction using a Doc-It imaging station, non-green colonies appeared after 24 h induction of 200 ng/mL ATc.