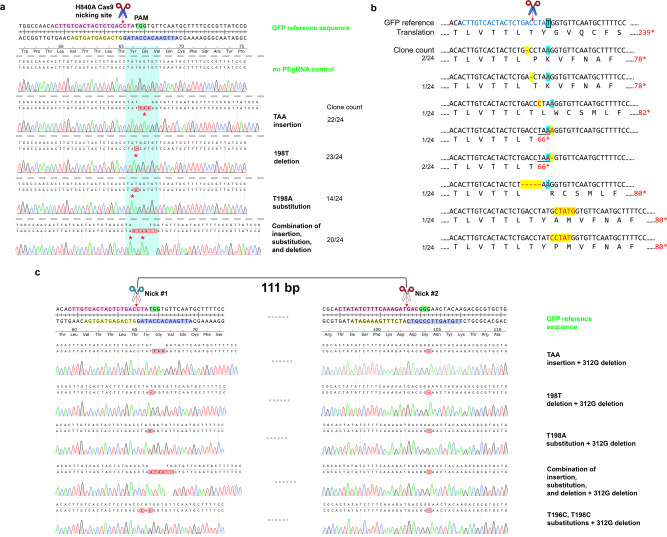
Fig. 2. Evaluation of plasmid-based editing by CRISPR-Prime Editing system using Sanger sequencing.
Stop codon is displayed as an asterisk under the nucleotide sequences, the potential Cas9 H840A nicking site is indicated by a red arrow. The 20 nt protospacer, the PAM sequence, the PBS, and the RTT are highlighted in pink, green, yellow, and blue, respectively; while the cyan masked Sanger sequencing traces show the sequence to be replaced by the RTT that will contain the different edits designed. a Twenty-four randomly picked colonies of each designed DNA engineering were Sanger sequenced and traces were aligned to the targeted locus of the GFP coding sequence. The correctly edited clone numbers and the total sequenced clone numbers are shown on the right of the figure. b Shown are the recorded target-specific unintended edits of the 1 bp substitution. The target T is boxed and masked in light blue. The GFP reference DNA sequence and the translated amino acid sequences are show on the top row. The corrected edited nucleotide is in red and masked with light blue, while the unexpected mutations (off-target) are in red and masked with yellow. The recorded off-target clone numbers and the total sequenced clone numbers are shown on the left of the figure. c Sanger sequencing traces of the successfully dual-edited clones by CRISPR-Prime Editing. Two nicks are 111 bp away, the left nick (nick #1) is introduced by pPEgRNA (ColE1 ori), and the right nick (nick #2) is introduced by pVRb_PEgRNA (pSC101 ori). The combinations of 3 bp insertion, 1 bp deletion, 1 bp substitution, 2 bp substitution, combinatory editing, and 1 bp deletion are displayed.