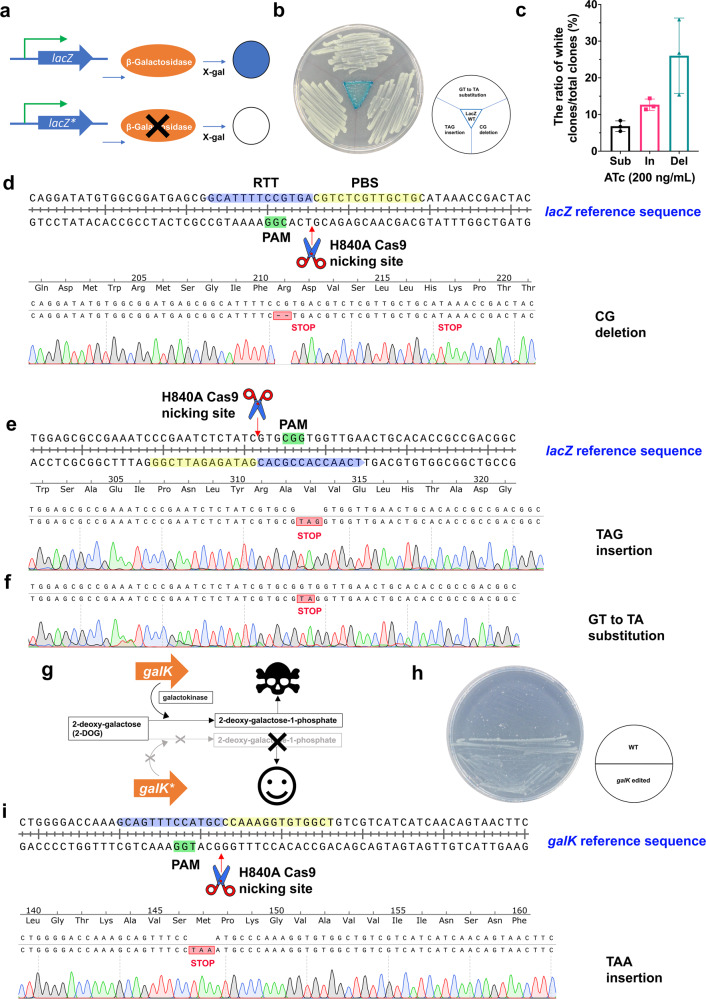
Fig. 4. CRISPR-Prime Editing for E. coli is capable of chromosomal DNA editing.
a A graphic illustration of the function of lacZ gene. The star within the gene box represents a stop codon being introduced. b Three clones of E. coli MG1655, where the inactivation of lacZ was confirmed by Sanger sequencing and a wild type E. coli MG1655 were re-streaked on an agar plate with X-gal. c A bar chart shows the editing efficiencies of chromosomal DNA engineering by 3 bp insertion, 2 bp deletion and 2-bp substitution by calculating the ratio of white clones/total clones on an induction LB plate with X-gal supplemented. Mean and standard deviation of three biological replicates are shown. d–f Eight non-blue colonies were picked and Sanger sequenced. Sequencing traces aligned with non-edited lacZ reference sequences are displayed. Shown are alignments of 2 bp deletion (d); 3 bp (stop codon TAG) insertion (e); and 2-bp substitution (f). g A graphic illustration of the function of galK gene. h An agar plate view of the successful galK gene inactivation in E. coli MG1655 strains by CRISPR-Prime Editing. i Four colonies from the 2-DOG supplemented plate were picked and Sanger sequenced. Sequencing traces aligned with non-edited galK reference sequences are displayed. The alignment of 3 bp (stop codon TAA) insertion is shown. The potential Cas9 H840A nicking site is indicated by a red arrow. The 20 nt protospacer, the PAM sequence, the PBS, and the RTT are highlighted in pink, green, yellow, and blue, respectively. The translated amino acid sequences together with the introduced stop codons are labeled underneath each nucleotide sequence. Numbers in red on the right side show the correct and total Sanger sequenced colonies.