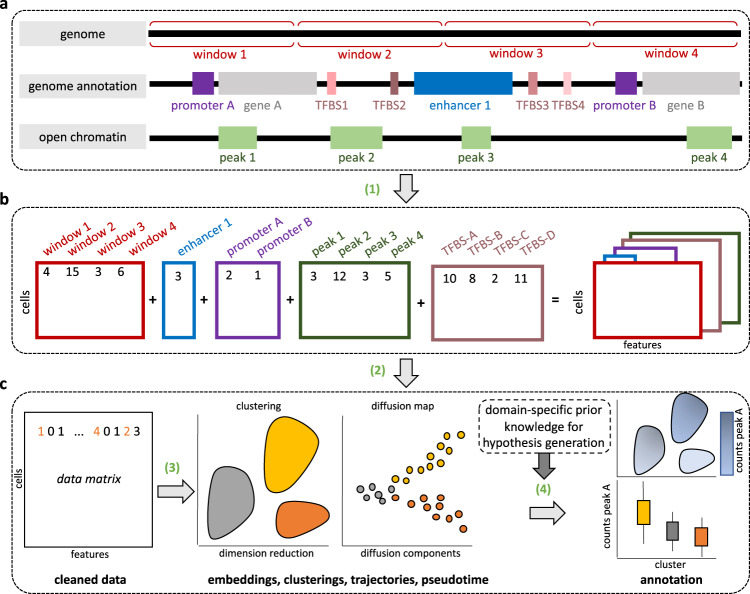
Fig. 1. EpiScanpy analysis workflow.
a epiScanpy quantifies chromatin openness and DNA methylation at different sets of genomic regions to b construct count matrices (1) with read counts (for scATAC-seq) or DNA methylation levels (for single-cell DNA methylation). c After data pre-processing (2), unsupervised learning algorithms (clusters, trajectories, lineage trees) are applied (3). Differential openness and methylation callings allow for identification of marker loci, which can be used for cell type and lineage tree identification (4).