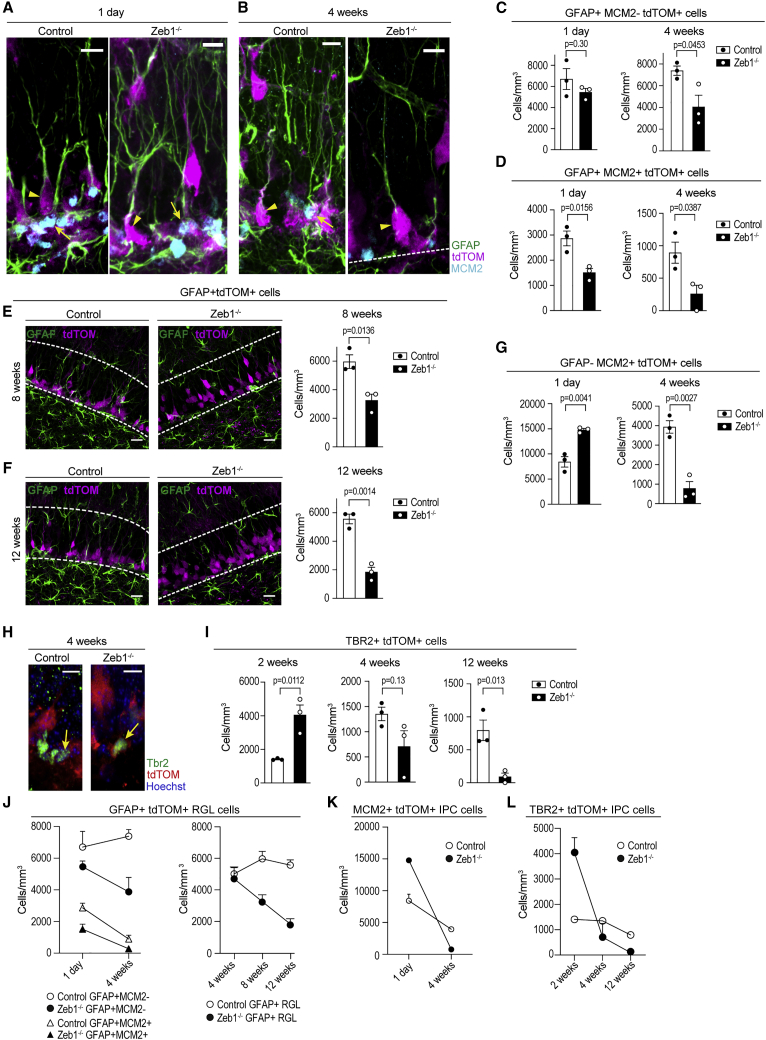
Figure 3.
Effects of Zeb1 loss in RGL cells and IPCs
(A and B) Representative images of quiescent (GFAP+MCM2−tdTOM+; arrowheads) and activated (GFAP+MCM2+tdTOM+; arrows) RGL cells in the SGZ of control and Zeb1−/− mice at 1 day (A) and 4 weeks (B) post-induction.
(C) Numbers of quiescent RGL cells at 1 day and 4 weeks post-Zeb1 deletion.
(D) Numbers of activated RGL cells at 1 day and 4 weeks post-Zeb1 deletion.
(E and F) Representative images and quantification of RGL cells at 8 (E) and 12 weeks (F) post-induction.
(G) Numbers of GFAP-MCM2+tdTOM+ IPCs at 1 day and 4 weeks post-Zeb1 deletion.
(H and I) Representative images at 4 weeks post-induction (H) and quantification of TBR2+ IPCs at 2, 4, and 12 weeks post-Zeb1 deletion (I).
(J–L) Summary graphs depict quiescent/activated (left) and total (right) RGL populations in control and Zeb1−/− mice (J). Summary graphs outline the temporal changes of GFAP−MCM2+ (K) and TBR2+ (L) IPCs.
Dots represent individual mice (minimum two sections analyzed per animal), except in (J)–(L), where dots represent averages. Numerical data are shown as mean ± SEM. Dashed lines in images demarcate DG boundaries. Scale bars: 20 μm (insets: 10 μm).